FJ, AMS: Department of Animal Science, Agriculture Experiment Station, University of Tennessee, Knoxville, TN 37996; AGM: Department of Animal Science, Brehm Bldg, 2505 River Drive, University of Tennessee, Knoxville, TN 37996; Tel: 856-974-3123; Fax: 865-974-7297; E-mail:amathew@utk.edu.
Mathew AG, Jackson F, Saxton AM. Effects of antibiotic regimens on resistance of Escherichia coli and Salmonella serovar Typhimurium in swine J Swine Health Prod. 2002;10(1):7-13. Available as a PDF
Original research
Peer reviewed
Effects of antibiotic regimens on resistance of Escherichia coli and Salmonella serovar Typhimurium in swine
Alan G. Mathew, MS, PhD; Felix Jackson, MS; Arnold M. Saxton, MS, PhD
Summary
Objective: To determine the effects of antibiotic dosing schemes on resistance patterns of naturally occurring Escherichia coli and a challenge strain of Salmonella serovar Typhimurium in swine.
Methods: In two trials, 96 weaned pigs were inoculated with Salmonella Typhimurium prior to being treated with antibiotics. Treatments included maximum label use, rotation of similar and non-similar antibiotics, increasing gradient doses, and pulse dosing, for a period of 2 weeks post challenge. Fecal samples were obtained before, during, and after antibiotic treatment for isolation of the challenge organism and E coli for resistance analysis using a minimum inhibitory concentration analysis.
Results: The E coli from pigs treated with pulse doses of antibiotics had lower resistance compared to other groups. Greater resistance occurred in E coli when similar antibiotics were used in rotation. Resistance of Salmonella Typhimurium was not affected by treatment and remained low throughout the study.
Implications: Antibiotic regimens appear to affect antibiotic resistance in bacteria associated with swine. Thus, dosing schemes that will simultaneously decrease risks posed by antibiotic resistance elements should be considered when formulating effective therapeutic or subtherapeutic antibiotic treatments. Such practices should address consumer concerns regarding agricultural use of antibiotics and increase acceptance of pork products.
Keywords: swine, antibiotic
resistance, Escherichia coli, Salmonella Typhimurium
swine, antibiotic
resistance, Escherichia coli, Salmonella Typhimurium
Received: January 16, 2001
Accepted: August 3, 2001
The practice of providing antibiotics to livestock is well established. Antibiotics are used in food animals therapeutically to treat disease, and sub-therapeutically to improve performance and to modify nutrient utilization. Subtherapeutic use of antibiotics such as sulfonamides, aminoglycosides, and tetracyclines has for many years represented the largest and most controversial category of antimicrobial use in animals.1 Some health experts believe that these feed additives are helping to establish or are contributing to a reservoir of drug resistant microorganisms which may be capable of transferring their drug resistance to animal or human pathogens.2,3
A number of earlier studies have demonstrated the development of drug resistance and in vivo resistance transfer between enteric bacteria, especially isolates from animals fed antibiotics.4-6 However, little information is available indicating how specific dosing schemes may affect antibiotic resistance patterns in bacteria. Thus, the objective of this research was to compare the effects of antimicrobial regimens on resistance patterns of Escherichia coli and Salmonella serovar Typhimurium in weaned pigs.
Materials and Methods
Study design
Phase One. Two replicate trials were conducted using a previously established Salmonella Typhimurium challenge model,7 with 48 weaned pigs that had not been treated with antimicrobials in each replicate trial. Pigs were derived from sows on the University of Tennessee Agricultural Experiment Station Blount Swine Unit, and sows had received no antibiotics within the previous year. Pigs were weaned at 18 to 21 days of age, blocked by litter, and divided into six groups of eight pigs each. Groups were then randomly assigned to treatment rooms. Pigs were challenged on day 2 post weaning via intranasal inoculation with 109 colony forming units (CFU) of Salmonella Typhimurium (Strain 798-4232, National Animal Disease Control, USDA, Ames, Iowa)8 derived from a confirmed case of swine salmonellosis. Delivery and concentration of the challenge organism dose were based on information from a previous study.8
Phase Two. In the second phase of the experiment, pigs from the first replicate trial of Phase One were maintained in the above facility in their respective treatment rooms throughout the finishing period. On day 30 post challenge, pigs from the second replicate trial of Phase One were moved to a separate facility that more closely simulated a modern swine growing-finishing facility, located approximately 97 km from the first research facility.
Housing and feeding
The isolation facility used for the first phase of the study was a newly constructed, intensive animal research facility where no pigs had been previously housed, located at the University of Tennessee Agriculture Experiment Station near the Knoxville campus. Each room was self-contained, with independent ventilation and waste removal systems to minimize the possibility of cross contamination of bacteria between rooms. Each room contained one nursery pen (1.486 m2), one finisher pen (5.95 m2), and associated feeding and water equipment. Floors were solid concrete. Manure was removed from the room on a daily basis. Biosecurity measures included use and changing of disposable boots and coveralls, and use of chlorhexidine diacetate disinfectant boot washes (Nolvasan; Fort Dodge, Overland Park, Kansas) before each room entry. Boot washes were changed every 3 days, or sooner if there was obvious accumulation of dirt, manure, or other contaminants. Rooms were provided 12 hours of light per day. Temperature in rooms was initially maintained at 28°C and was decreased as the pigs aged, according to recommendations outlined in the Pork Industry Handbook.9 Pigs were provided ad libitum access to water and feed throughout the experiment.
The facility used for the second phase of the experiment provided 5.77 m2 of pen floor space, with approximately half of the floor consisting of solid concrete and half consisting of concrete slats. The room was ventilated passively via curtain sides. Although pigs were penned by treatment group, all pigs were housed in a common room, with groups separated by one empty pen.
An antibiotic-free diet, containing 22.3% crude protein, 1.3% lysine, and 8.1% fat, and meeting or exceeding NRC10 recommendations for other nutrients, was provided during the first 21 days post challenge (including the entire antibiotic treatment period). A phase two diet, providing 19.8% crude protein, 1.1% lysine, and 5% fat, and meeting or exceeding NRC10 recommendations for other nutrients, was provided for 20 days immediately following the antibiotic treatment period. Subsequent grower and finisher diets were typical of those used in the industry and were based on NRC recommendations.
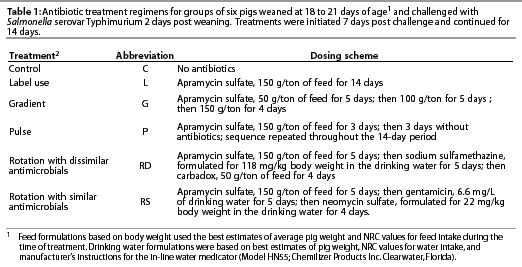
Antibiotic treatments
The antibiotic treatments, described in Table 1, were initiated on Day 7 post challenge (9 days post weaning) and continued for 14 days.
Challenge inoculum preparation
Twenty µL of stock solution containing the Salmonella Typhimurium challenge strain was inoculated into 200 mL of Luria Broth, consisting of 10 g per L Bacto-tryptone, (DIFCO, Detroit, Michigan), 5 g per L yeast extract (DIFCO), and 10 g per L sodium chloride, and adjusted to pH 7.5 with sodium hydroxide. The inoculated broth was incubated aerobically overnight at 37°C, and the resulting culture was used for the challenge as described above.
Isolation of non-pathogenic E coli from rectal swabs
Rectal swabs were collected from all pigs on Days 3, 6, 10, 13, 17, 31, 41, 70, and 101 post challenge using Dacron fiber tipped swabs (Fisher, Houston, Texas). The samples were then placed in sterile test tubes, transported on ice to the laboratory, inoculated onto Lactose MacConkey Agar (DIFCO), and incubated aerobically overnight at 37°C for the isolation of coliforms. Colonies with the pink coloration and morphology of E coli were subjected to an API20E test (BioMerieux Vitek, Inc, Hazelwood, Missouri) for confirmation to species. Confirmed E coli were then inoculated onto Trypticase Soy Agar with 5% sheep blood (BBL, Cockeysville, Maryland) for determination of hemolysis activity. Only non-hemolytic E coli were used for resistance testing.
Isolation of salmonellae
After the swabs had been inoculated onto media for E coli recovery, the tips were removed and placed in glass tubes to which were added 5 mL of a cryomedium containing 100 mL sterile water, 3 g trypticase soy (DIFCO), 14 mL horse serum (Gibco, Rockville, Maryland), and 28 mL glycerol (Fisher). The tubes were then briefly vortexed (S/P Vortex Mixer; VWR Scientific Products, San Dumas, California) and 2.5 mL of suspension was pipetted into a Stomacher bag (Seward, London, UK) containing 80 to 100 mL of tetrathionate broth (DIFCO). The Stomacher bags were then sealed and incubated aerobically at 42°C for 18 to 24 hours.
After enrichment in tetrathionate broth, cultures were plated on XLT4 medium (DIFCO) to which nalidixic acid had been added (after cooling to 50°C) to a concentration of 50 µg per mL to exclude the growth of salmonellae other than the challenge isolate. Plates were incubated aerobically at 37°C for 18 to 24 hours and resulting colonies were biochemically tested using Triple Sugar Iron (DIFCO) and Lysine Iron Agar (DIFCO) slants. Slants were incubated at 37°C for 24 hours and color changes were assessed to confirm isolates as salmonella.11
Antibiotic sensitivity testing
A maximum of four confirmed isolates of E coli and four of Salmonella Typhimurium were obtained from each pig sample. Individual colonies were transferred to sterile test tubes containing 5 mL Mueller-Hinton II broth (BBL). Inoculated tubes were placed in racks at a 45 degrees angle in a shaking water bath, and incubated aerobically at 37 degrees C. Bacterial cultures were grown to a concentration matching a 0.5 McFarland standard turbidity level (5 to 8 hours), resulting in a concentration of approximately 108 CFU per mL.12 A 1:10 dilution of bacterial suspension was then made in sterile water, 5 µL of the dilution was added to 45 µL of Mueller-Hinton II broth, and this suspension was added to a well in a microtiter plate containing 50 µL of antibiotic diluted in Mueller-Hinton broth. The resulting bacterial-antibiotic suspension in each well then contained approximately 5 x 105 CFU per mL.
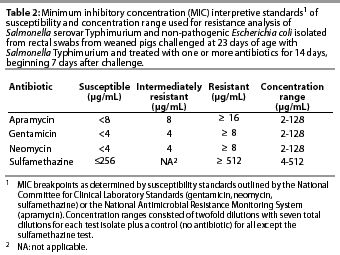
The antibiotics used in the susceptibility test included gentamicin (ICN Biochemical Inc, Aurora, Ohio); apramycin (Eli Lilly and Company, Indianapolis, Indiana); neomycin (Sigma, St Louis, Missouri); and sulfamethazine (Sigma). Twofold dilutions of each antibiotic were made in 96-well microtiter plates over the range of concentrations specific to that drug (Table 2). The dilution range was based on clinical resistance breakpoints12 and preliminary work in our laboratory to determine the most common minimum inhibitory concentrations (MIC) among swine-derived enteric bacteria. Each microtiter plate enabled the testing of 11 test isolates over eight dilutions, and a control isolate (E coli strain 25922, American Type Culture Collection, Rockville, Maryland) previously determined to be sensitive to the test drugs. Microtiter plates were incubated aerobically overnight at 37°C, and MICs were determined by inspection of wells for growth. The MIC was determined from the first well of each dilution column in which no growth was noted. Determination of clinical resistance was based on previously established breakpoints according to the National Committee for Clinical Laboratory Standards12 (neomycin, gentamicin, sulfamethazine) or the National Antimicrobial Resistance Monitoring System13 (apramycin), as indicated in Table 2.
Statistical analysis
For Phase One of the experiment (up to Day 31 post challenge), a randomized block design, blocked on two replicates, with repeated measures, was used to analyze data pertaining to antibiotic resistance.14 Sensitivity breakpoint data were linearized by a log base 2 transformation and comparisons of MICs were made using least squares means. Differences among all means were tested with Fisher's Protected LSD. Differences were considered significant at P<.05. The experimental units for antibiotic treatments were pens. The model was
![]()
Data from Phase Two (Days 41 to 101 post challenge) were analyzed using a similar model, except barn condition (Rep) was a fixed effect, comparing effects of housing groups in isolation versus housing groups in a common facility. Statistical replication in the whole plot was obtained by using pig as the experimental unit in a completely randomized design.
Results
E coli analysis
Throughout the study, E coli were isolated from nearly all swabs. Treatment effects were noted (P<.0001) for resistance to apramycin (Table 3), gentamicin (Table 4), neomycin (Table 5), and sulfamethazine (Table 6) in E coli isolates during the first phase of the experiment. Interactions between treatment and Day post challenge were noted for E coli (P<.0001), with the greatest resistance generally occurring during or immediately after antibiotic treatment.

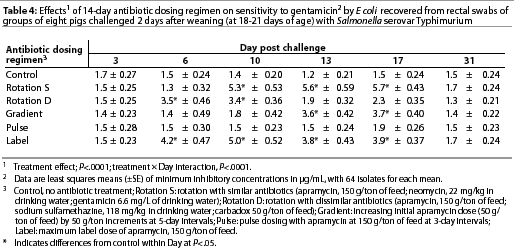


The RS treatment (Table 1) produced isolates with the greatest resistance to apramycin, neomycin, and gentamicin. Increased resistance to apramycin developed by Day 10 post challenge and was maintained through Day 17 post challenge (Table 3). A similar trend was observed for all antibiotics except sulfamethazine.
Among test regimens, pulse dosing with apramycin produced the least resistance to apramycin. With the exception of Day 17 post challenge, MICs of the pulse dose group did not differ statistically from those of the control group, which did not receive antibiotics.
The RD treatment (Table 1) produced the greatest MICs to sulfamethazine. An increase occurred by Day 10 and MICs decreased by Day 31 (Table 6). Increased MICs for sulfamethazine were also noted for the RS treatment (in which sulfamethazine was not used). However, all E coli isolates from all treatment groups remained clinically susceptible to sulfamethazine (MIC<256 µg per mL).
During the second phase of the study, Day 41, sensitivity to apramycin of E coli isolated from the RS treatment group differed (P<.05) between groups housed in isolation and in common housing (data not shown). On that day, MICs were greater in isolates from pigs housed in isolated rooms (15.5 µg per mL) than in isolates from pigs housed in the common growing-finishing facility (2.4 µg per mL). Otherwise, for both housing treatments, MICs did not meet or exceed 8 µg per mL for apramycin (clinical resistance). A similar trend was noted for neomycin (data not shown). For both the RS and RD treatments, greater MICs to sulfamethazine were found in isolates from pigs in the common facility than in isolates from pigs in isolation (data not shown); however, MICs did not exceed the threshold for clinical resistance.
Salmonella Typhimurium analysis
The challenge organism was isolated in varying numbers from most pigs for up to 8 weeks post inoculation. Treatments did not affect sensitivity of Salmonella Typhimurium during either phase of the study (data not shown). Resistance to antibiotics remained low, and MICs did not exceed clinical resistance levels in any treatment group, not meeting or exceeding 4 µg per mL for the aminoglycosides or exceeding 256 µg per mL for sulfamethazine. The RD treatment was associated with the greatest MICs to sulfamethazine, as it was for E coli isolates, although MICs did not meet or exceed the threshold for clinical resistance.
Discussion
A total of 3428 isolates of non-pathogenic E coli and 335 isolates of Salmonella Typhimurium were tested with apramycin, gentamicin, neomycin, and sulfamethazine to determine the effects of antimicrobial regimens on resistance patterns. We also tested for resistance to carbadox using an anaerobic culture analysis based on the recommendation of the manufacturer of Mecadox (Pfizer Animal Health, Lee's Summit, Missouri.). This procedure results in MICs that are considerably lower than those observed with aerobic culture, and thus the data cannot be compared with traditional breakpoints. We observed no differences between treatments with regard to carbadox resistance (data not shown).
While pigs had not been exposed to antibiotics previous to the study, and sows had not received antibiotics for at least 1 year prior to the study, some sows may have been exposed to antibiotics before entering the breeding herd. Such earlier exposure may have affected patterns of bacterial isolates in subsequent litters; however, because pigs were blocked by litter prior to allocation to treatments, any effect of previous sow exposure to antibiotics should have been consistent across treatments.
In this study, pulse dosing with apramycin was associated with the least resistance in E coli, and was comparable to the control, for which antibiotics were excluded. The rotation with similar antibiotics (apramycin, gentamicin, and neomycin) was associated with the greatest MICs for E coli, and isolates were commonly resistant to all three aminoglycosides in this treatment. This is in agreement with an earlier study15 in which we observed that apramycin and neomycin resistance usually occurred as part of a multiple resistance pattern. Although the mechanisms resulting in cross resistance and other multiple resistance patterns are complex, it may be concluded from this study and others7,16 that use of drug combinations may result in multiple resistance after only a few days of treatment. Maximum label use of apramycin was also associated with greater resistance to apramycin, gentamicin, and neomycin, compared to controls, pulse dosing, and rotation with dissimilar antibiotics. We also observed an increase in resistance as the concentration of apramycin increased in the gradient dose treatment. That resistance occurred later than with the label dose treatment but was expressed by higher MICs. Minimum inhibitory concentrations declined subsequent to withdrawal of the antibiotic from the feed for all regimens except the control, in agreement with earlier studies15,17 in which we also noted a decline in resistance to apramycin following withdrawal of the drug. This pattern is in contrast with what has been observed for tetracyclines,18, 19 suggesting that resistance patterns are antibiotic-specific.
When data for each treatment were pooled over all sampling days, the greatest MICs in E coli to sulfamethazine were associated with rotation with similar antibiotics (127.9 µg per mL) and dissimilar antibiotics (131.1 µg per mL). Minimum inhibitory concentrations for sulfamethazine were inherently greater than for other antibiotics used in this study, and thus, while numerically greater compared to those for apramycin, gentamicin, and neomycin, these MICs do not indicate greater clinical resistance to sulfonamides.
In the second phase of this study, we also compared effects of isolation versus common housing of treatment groups. Most data were similar for both housing treatments; however, for the RS treatment, isolates of E coli from pigs maintained in isolation had greater (P<.05) MICs for apramycin on Day 41 post inoculation (15.5 µg per mL) compared to pigs held in the common facility (2.4 µg per mL). A similar increase (P<.05) was noted in resistance to neomycin by those isolates (data not shown). This may indicate that a more diverse microflora present in communally housed animals may promote a loss of resistance in the absence of antibiotic treatment. It might also be noted that pigs that were housed in isolated rooms were on solid cement flooring, whereas the flooring in the common facility consisted of partial concrete slats, which may have allowed less contact with manure. Langlois et al19 observed that housing conditions in intensive pig units, which allow high fecal contact, are probably important factors to consider when treating animals for infection. However, half of the floor in our common facility, a significant portion, was solid, and probably still allowed significant contact between pigs and manure, similar to pens in the isolation facility.
In evaluating Salmonella Typhimurium, it was postulated that low resistance of this organism to the test antibiotics was associated with housing the animals in a new facility, where no pigs had been previously housed. This is in contrast to our earlier study, which took place in a finishing unit built in the 1970's and used frequently thereafter.7 In that study, the same Salmonella Typhimurium challenge strain (delivered at the same dose) showed increased resistance to apramycin subsequent to treatment with apramycin for 14 days. This may indicate that resistance genes from other salmonellae were pre-existing in that building (earlier groups had occasionally been diagnosed with salmonellosis), or similar genes may have occurred in other bacteria resident to the building. It thus appears that E coli may have a greater ability to mutate to resistance or acquire pre-existing resistance elements in new or cleaner facilities, compared to Salmonella. However, it should also be noted that our recovery of salmonella was strictly selective for the challenge strain, which was known to be initially sensitive to the test antibiotics. This is in contrast to E coli, for which a variety of isotypes were likely obtained, including some strains which may have been resistant initially, and which may have then proliferated or disseminated resistance genes within species upon exposure to antibiotics.
Implications
- Antibiotic dosing regimens have a significant effect on antibiotic resistance patterns in E coli associated with swine.
- Pulse dosing limits the prevalence of resistant E coli among dosing schemes tested, whereas increasing doses of apramycin promote greater resistance.
- Rotations using aminoglycosides promote greater resistance to selected antibiotics in that family of anti-microbials.
- Resident E coli appear to acquire antibiotic resistance more easily than Salmonella Typhimurium.
- Housing treated and untreated pigs together in common facilities does not appear to promote resistant organisms in untreated pigs, and may help to reduce the numbers of resistant isolates in pigs that have been treated with antibiotics.
Acknowledgement
This work was funded in part by the National Pork Producer's Council, on behalf of the National Pork Board.
References -- refereed
1. National Research Council. The use of drugs in food animals: benefits and risks. Washington, DC: National Academy Press; 1999:75-86.
2. Tollefson L, Fedorka-Cray PJ, Angulo FJ. Public health aspects of antibiotic resistance monitoring in the USA. Acta Vet Scand Suppl. 1999;92:67-75.
3. van den Bogaard AE, Stobberingh EE. Antibiotic usage in animals: impact on bacterial resistance and public health. Drugs. 1999;58:589-607.
4. Mitsuhashi S, Herada K, Kameda M. On the drug resistance of enteric bacteria. Spontaneous and artificial elimination of transferable drug resistance factors. Jpn J Microbiol. 1961;31:119-122.
5. Watanabe T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963;27:87.
6. Smith HW. Persistence of tetracycline resistance in pig E coli. Nature. 1975;258:628-630.
7. Ebner PD, Mathew AG. Effects of antibiotic regimens on fecal shedding patterns of pigs infected with Salmonella typhimurium. J Food Protect. 2000;63:709-714.
8. Fedorka-Cray PJ, Collins Kelley L, Stabel GJ, Gray JT, Laufer JA. Alternate routes of invasion may affect pathogenesis of Salmonella typhimurium in swine. Infect Immun. 1995;63:2658-2664.
9. Pork Industry Handbook. Lafayette Indiana: State Cooperative Extension Service, National Pork Producers Council, and United States Dept of Agriculture; 1996.
10. Nutrient Requirements of Swine. 10th ed. Washington, DC: National Research Council; 1998.
11. Bacteriological Analytical Manual/Food and Drug Administration. 8th ed. Gaithersburg, Maryland: Association of Official Analytical Chemists International; 1998.
12. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Tentative Standard. Villanova, Pennsylvania: National Committee for Clinical Laboratory Standards; 1997.
13. National Antimicrobial Resistance Monitoring System - Enteric Bacteria. Washington, DC: Food and Drug Administration, United States Dept of Agriculture, and Centers for Disease Control and Prevention; 2000.
14. SAS SAS/STAT Users Guide (Version 6). 1st ed. Cary, NC: SAS Institute Incorporated; 1990.
15. Mathew AG, Saxton AM, Upchurch WG, Chattin SE. Multiple antibiotic resistance patterns of Escherichia coli isolated from swine farms. Appl Environ Microbiol. 1999;65:2770-2772.
17. Mathew AG, Upchurch GW, Chattin SE. Incidence of antibiotic resistance in fecal Escherichia coli isolated from commercial swine farms. J Anim Sci. 1998;76:429-434.
18. Langlois BE, Cromwell GL, Stahly TS, Dawson KA, Hays VW. Antibiotic resistance of fecal coliforms after long-term withdrawal of therapeutic and subtherapeutic antibiotic use in a swine herd. Appl Environ Microbiol. 1983;46:1433-1434.
19. Langlois BE, Dawson KA, Leak I, Aaron KD. Effects of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic exposed herd. Appl Environ Microbiol. 1988;50:1341-1344.
References -- Non refereed
16. Cullen P, Mathew AG, Clift R, Chattin SE. Effects of management and environmental conditions on antibiotic resistance in bacteria associated with swine. Proc 101st Meet Am Soc Microbiol. Orlando, Florida. 2001:218.
