Original research
Peer reviewed
Establishment of a herd negative for porcine reproductive and respiratory syndrome virus (PRRSV) from PRRSV-positive sources
Montserrat Torremorell, DVM, PhD; Camille Moore, DVM; William T. Christianson, DVM, PhD
MT, WTC: Pig Improvement Company, Franklin, KY 42134 CM: St-Césaire, Quebec, Canada. Corresponding author: Montserrat Torremorell, 3033 Nashville Rd, Franklin, KY 42134;
Tel: 270-586-9224; Fax: 270-586-0312; E-mail: montse.torremorell@pic.com.
Torremorrell M, Moore C, Christianson WT. Establishment of a herd negative for porcine reproductive and respiratory syndrome virus (PRRSV) from PRRSV-positive sources. J Swine Health Prod. 2002;10(4):153-160. Also available as a PDF.
Summary
Objective: To establish a herd negative for porcine reproductive and respiratory syndrome virus (PRRSV) from PRRSV-positive sources by management of the gilt pool and batching of pig flow.
Methods: Two groups of PRRSV-positive gilts were housed in common acclimatization areas for 70 to 100 days, bred in an off-site finishing facility, and farrowed in a separate facility. Piglets weaned at 5 to 7 days of age were moved to off-site nurseries in weekly batches and mixed with cohort sentinel piglets from a PRRSV-negative herd. Each nursery batch was tested for PRRSV twice by PCR and once by ELISA. Two to five negative batches were grouped in off-site grower facilities, and tested again. Negative groups moved to a quarantine facility. Negative quarantine groups were used for stocking the PRRSV-negative herd.
Results: Of the 31 batches of nursery pigs produced, three batches, born 2, 4, and 6 weeks after farrowings started, were PRRSV-positive. Six grower groups (comprising 23 nursery batches) that were PRRSV-negative after repeated testing were assembled into five groups in a quarantine facility and remained PRRSV-negative. Two grower groups were rejected. A total of 9500 pigs were produced, from which 3415 pigs were selected to stock the PRRSV-negative herd.
Implications: A PRRSV-negative population was established from positive sources by managing the gilt pool and batching the pig flow, allowing for preservation of elite genetics. It appeared that PRRSV infection, indicated by lack of seroconversion in the offspring, eventually either disappeared or became inactive in the donor gilt population.
Keywords: swine, porcine reproductive and respiratory syndrome, gilt acclimatization, porcine reproductive and respiratory syndrome virus eradication, false positive
swine, porcine reproductive and respiratory syndrome, gilt acclimatization, porcine reproductive and respiratory syndrome virus eradication, false positive
Received: September 28, 2001
Accepted: March 18, 2002
Porcine reproductive and respiratory syndrome (PRRS) was first de-scribed in the United States in 19871 and the virus (PRRSV) was isolated in Europe in 1991.2 Since then, research efforts have been directed towards understanding the pathogenicity of the virus and controlling the clinical and economic impact of the disease. Among strategies to control virus infection, isowean and segregated early weaning (SEW) have been proposed as alternatives to produce PRRSV-negative pigs from PRRSV-positive sources.3 However, results have not always proven successful, as consistent production of batches of negative pigs depends on the infection status of the sow herd and prevention of infection to the offspring and also laterally to other pigs in the batch and to other batches.4
Animals may remain infected for prolonged periods. Published data indicate that experimentally infected nursery age pigs may harbor PRRSV in the tonsils for up to 157 days5 and in one study, transmitted infection to contact controls for up to 62 days post infection.6 In another study, experimentally infected breeding age animals harbored the virus for up to 86 days post infection.7 Eventually, a protective immune response develops and the virus is cleared from the pig's body, which in some cases may take up to 5 months.8 It has been reported that PRRSV infection sporadically disappears from small herds upon interruption of gilt introduction,9 and that it is possible to eradicate PRRSV from infected herds.10,11
This study reports the establishment of a PRRSV-negative sow herd from PRRSV-positive sources in order to preserve the elite genetics of the donor herds. The approach was novel in its use of a discrete population of donor animals from two sources commingled in acclimatization areas for a minimum of 4 months before breeding was initiated. No additional groups of gilts were added to the original donor population. In addition, managing the PRRSV-negative offspring in weekly nursery batches housed in separate facilities, and extensive testing as the batches moved through the grower and quarantine facilities, contributed to the success of the project.
Material and methods
Gilt sources
Two PRRSV-positive herds (A and B) were used as donors of PRRSV-positive gilts. In both herds, recurrent PRRSV infections occurred in nursery and grower pigs, with circulating virus demonstrated by virus isolation. Monthly testing of 30 animals showed that all pigs were seropositive by 4 months of age (HerdChek ELISA; IDEXX Laboratories, Westbrook, Maine). These herds contained genetic lines unique to the production system; therefore, the possibility of stocking the new herd from a PRRSV-negative source was not an option.
Study design
Acclimatization of PRRSV-positive donor gilts. Two groups of gilts from Herd A and two groups from Herd B were commingled at 2 to 3.5 months of age in two finishing locations (Acclimatization Areas C and D) in order to enhance consistent exposure to herd-specific microflora, particularly to different strains of PRRSV known to be endemic in each herd. Animal movement is shown in Figure 1. The first group of animals, consisting of 500 gilts originating from herd A and 600 gilts from herd B, was delivered to Acclimatization Area C, and 8 weeks later, the second group, also consisting of 500 gilts from herd A and 600 gilts from herd B, was delivered to Acclimatization Area D. After 70 days for Acclimatization Area C and 100 days for Acclimatization Area D, sera from all gilts were tested for PRRSV by polymerase chain reaction (PCR). Gilts that were PRRSV-positive were removed from the group. The remaining gilts were held for a further 30 days before breeding.
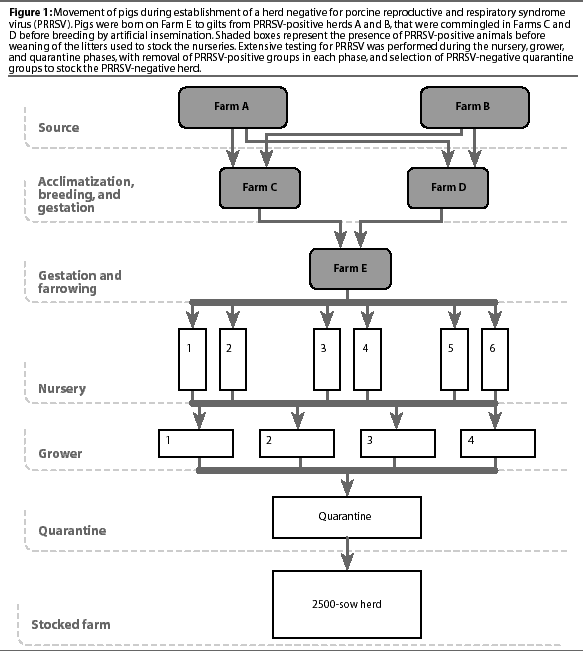
Breeding, gestation, and farrowing of donor gilts. At approximately 210 days of age, gilts in the two finishing locations were bred by AI and pregnancy-checked 30 days later. Gilts were housed in gestation pens for 11 weeks. Five weeks before farrowing, gilts were moved to crates in a gestation and farrowing facility (Farm E). Three farrowing rooms were used alternately and managed all in-all out on a weekly basis. Gilts were moved into a farrowing room 3 days before their due date. Piglets were weaned at 5 to 7 days of age and moved to off-site, isolated nurseries located approximately 35 km from the farrowing farm and at least 15 km from the closest acclimatization facility (Acclimatization Area C). To prevent infection with targeted microorganisms endemic in the herds,12 piglets were medicated with antibiotics intramuscularly for 2 days before and on the day of weaning, and in the drinking water for 1 week postweaning.
Producing PRRSV-negative nursery pigs. Six portable nursery trailers (Double-L, Monona, Iowa) were used as nurseries, with two trailers in each of three locations. Each trailer had a capacity of 300 to 350 piglets, 24 pens with 12 to 14 pigs per pen, and open gating allowing nose-to-nose contact between pens. Trailers were managed all in-all out on a weekly basis. A batch was defined as the weekly pig production housed in one nursery trailer. Age-matched sentinel pigs, weaned at 5 to 7 days from a PRRSV-seronegative herd (tested monthly by HerdChek ELISA), were mixed with the principal pigs at a ratio of 1:14 (24 sentinel pigs in each batch, one per pen).
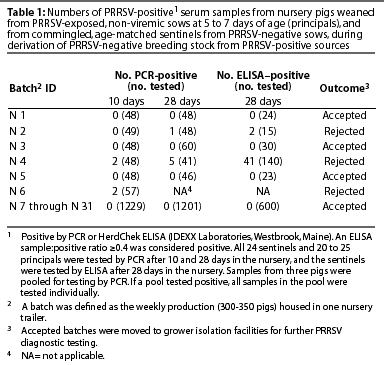
Ten days after weaned pigs entered the nursery, blood samples collected from all sentinel pigs and 20 to 25 randomly selected principal pigs were tested for PRRSV by PCR. After 4 weeks in the nursery, blood samples from all sentinel pigs and from 20 to 25 randomly selected principals were again tested by PCR, and samples from sentinels were also tested by HerdChek ELISA. The principal pigs were expected to be seropositive because of maternal antibodies and were not tested by ELISA.
After 5 weeks in the nursery, PRRSV-negative batches were moved to off-site grower facilities, and PRRSV-positive batches were removed from the project and sold as feeder pigs. At the end of the nursery period, three to five sentinel pigs from each batch were euthanized, and tissues were submitted to the University of Minnesota Diagnostic Laboratory, St Paul, Minnesota, for PRRSV diagnostic testing.
Producing PRRSV-negative grower pigs. Four grower sites were alternated to accommodate pig flow. A grower group contained two to five batches of nursery pigs depending on site capacity. Nursery batches were moved into the grower site weekly until the site was full. Pen integrity from the nurseries was not maintained, and males and females were housed separately. Seven groups of grower pigs were assembled from the PRRSV-negative nursery batches. Two grower groups (Groups 2 and 4) were divided into subgroups A and B, because they occupied separate buildings in the grower facility.
Three weeks after each grower facility was totally populated, blood was collected from all pigs for testing by HerdChek ELISA. If all animals in a group were seronegative when initially tested, the group was selected for shipment to the quarantine facility.
A percentage of ELISA false-positive results was expected as noted by the manufacturer,13 and the positive samples were retested, using a different test, to confirm the true status of the sample. In Groups 1 through 5, PCR was the secondary test used on the ELISA-positive samples. In Groups 6 and 7, indirect fluorescent antibody (IFA) was the secondary test. Sero-positive animals were also necropsied and tissues were submitted for diagnostic testing for PRRSV (PCR, histopathology, and virus isolation). Two weeks after the sero-positive animals had been necropsied, blood samples were collected from ten in-contact animals and tested by ELISA. A grower group was considered negative if the secondary tests on the ELISA-positive samples, the tests on the necropsied animals, and the tests on the in-contact animals were all negative.
After the grower groups were designated negative, individual pigs were selected on the basis of genetics and shipped within 4 weeks to a quarantine facility to accommodate regulatory testing as well as the final PRRSV testing before the animals were used for stocking the new PRRSV-negative herd.
In groups with conflicting PRRS ELISA results, additional testing, including bioassay, was performed before groups were moved into the quarantine facility. Grower groups designated PRRSV-positive were removed from the project and sold as feeder pigs.
At the end of the 6- to 10-week growing period, slaughter checks were conducted on 30 of the remaining sentinels in each PRRSV-negative grower group.
Quarantine testing of PRRSV-negative pigs. Grower groups that tested PRRSV-negative were reassembled into quarantine groups. Grower Groups 1, 2B, 3, and 7 became quarantine Groups 1, 2B, 3, and 7, and quarantine Group 4-5 was assembled from grower subgroups 4A and 4B and grower Group 5. The quarantine facility was managed all in-all out by group. Groups remained in the quarantine facility for a minimum of 4 weeks. All pigs were tested for regulatory purposes within 1 week of arrival, and were tested at the same time by PRRS ELISA. In each group, a subgroup of 60 randomly selected animals was tested again 2 weeks later (3 weeks post entry into the quarantine facility), with the objective of detecting at least one positive sample, assuming at least a 5% prevalence of PRRSV with a 95% confidence interval.
Quarantine groups were selected for stocking the new PRRSV-negative herd if all animals were seronegative 1 week post entry into the quarantine facility, and if the subgroup of retested animals was also negative 3 weeks post entry. Samples from these groups with ELISA sample:positive (S:P) ratios >=0.4 were retested by PCR (Group 1) or by IFA (Groups 2 through 7). If these samples subsequently tested negative by either PCR or IFA, the group was considered suitable for stocking the PRRSV-negative sow herd, and was moved into the new farm, where a monitoring program, based on monthly representative testing of 30 blood samples by HerdCHek ELISA, was implemented.
Semen source
In order to prevent genetic lag throughout the project, elite boars were selected at 5 months of age from Herds A and B and blood tested by PCR within 2 weeks post selection. Samples of semen from all boars were individually tested by PCR at collection.
PCR testing
Serum samples from gilts and boars and semen samples from boars were submitted to Centro de Investigación en Salud Animal, Madrid, Spain, for testing using the RT-one-step PCR.14 Semen samples were individually tested. Serum samples were pooled in groups of three. If a pool tested positive, all samples in the pool were tested individually.
ELISA and indirect fluorescent antibody (IFA) testing
Serum samples were tested for PRRSV antibodies by HerdChek ELISA (IDEXX). Samples with ELISA S:P ratio >= 0.4 were considered positive. Beginning with the 15th batch, positive samples were retested by IFA as described by Yoon,15 with titers >=16 considered positive.
Necropsy
At the end of the nursery period, necropsies were performed on three to five sentinel pigs from each batch, and tissues were submitted to the University of Minnesota Diagnostic Laboratory (St Paul, Minnesota) for PRRSV diagnostic tests, including virus isolation (VI), PCR, histopathological examination, and immunohistochemistry (IHC).
In addition, at the end of the grower period, 30 randomly selected sentinel pigs were submitted from each group for slaughter check evaluation, consisting of visual inspection of the lungs, livers, skin, and snouts, and tissues with gross lesions were submitted to the University of Minnesota Diagnostic Laboratory for PRRSV diagnostic testing (VI and histopathology).
Necropsies were also performed on pigs that died suddenly, acutely sick pigs, and pigs suspected to be false-positive on the ELISA and IFA tests, and tissues were submitted to the University of Minnesota Diagnostic Laboratory for PRRSV testing as described above.
Bioassay
Tissue samples from individual pigs from grower Group 6 with conflicting PRRSV ELISA results were subjected to the swine bioassay technique to identify viable PRRSV.16 Tissues (sera, tonsils, lung, and lymph nodes) were harvested from 15 pigs and included in the homogenate for bioassay inoculation. Ten 3-week-old piglets from a PRRSV-negative source were inoculated intraperitoneally with the tissue homogenate, and blood samples collected from each pig weekly for 5 weeks were tested by HerdChek ELISA.
Results
Seropositive donor gilts
In the first group of 1100 donor gilts (Acclimatization Area C), 11 gilts tested by PCR 70 days after entering the unit were PRRSV-positive. These gilts were removed from the breeding group.
In the second group of 1100 donor gilts (Acclimatization Area D), all gilts tested by PCR 100 days after entering the unit were PRRSV-negative.
After removal of the PCR-positive gilts, both gilt populations (in Acclimatization Areas C and D) were considered exposed and non-viremic.
Nursery pigs
Among the 31 batches of nursery pigs (9500 pigs) produced, three batches were PRRSV-positive by PCR (Table 1). Two of these were positive when tested 10 days post entry into the nursery, and the third was positive when tested 28 days post entry. The three PRRSV-positive batches were from litters born 2, 4, and 6 weeks after farrowing started. All nursery batches born after that were PRRSV-negative.
The 28 batches of nursery pigs that were PRRSV-negative by PCR and ELISA were moved into the grower facilities (a total of 8960 pigs).
Grower pigs
Table 2 shows the results of testing by ELISA, PCR, and IFA in the grower animals. A total of seven grower groups were assembled from the 28 batches of PRRSV-negative nursery pigs. After testing, six grower groups were considered PRRSV-negative and were selected for stocking the new PRSSV-negative herd. In Groups 1, 2B, 3, 4 (A and B), 5, and 7, 39 of the 6042 samples tested were ELISA-positive (0.6%). Secondary testing by PCR was performed on 34 ELISA-positive samples from Groups 1 through 5, and secondary testing by IFA was performed on five ELISA-positive samples from Group 7. In addition, tissue submitted from the ELISA-positive animals was PRRSV-negative by PCR, VI, and IHC.

Rejected grower groups
Two groups of pigs in the grower stage, groups 2A and 6, comprising seven nursery batches, were rejected (Table 2).
Group 2A. When Group 2A was tested 5 weeks post placement into the grower facility, 12 ELISA-positive pigs were clustered in four of the 20 grower pens. An active infection was suspected, and the pigs housed in the half of the room containing the infected pens were slaughtered. Pigs in the other half of the room were moved to an off-site finishing facility, and infection in this group of 300 pigs was confirmed later: 17 samples were PCR-positive and 30 were ELISA-positive.
Group 6. Group 6 was rejected at the grower level because PRRSV serological results were unclear (Table 3). When the group was first tested, 3 weeks after entering the grower facility, nine of the 883 pigs were ELISA-positive. When these nine samples, and in addition, samples from all pigs with ELISA S:P ratios > 0.25 (a total of 50 samples) were tested by IFA, 12 pigs were IFA-positive. When the same 50 animals were retested 17 days later, five were ELISA-positive, and nine were IFA-positive. Fifteen pigs from this group were euthanized, and tissues were tested by bioassay. Bioassay tests conducted in ten 3-week old piglets were negative (ie, 5 weeks post inoculation, ELISA tests on all ten inoculated pigs were negative).
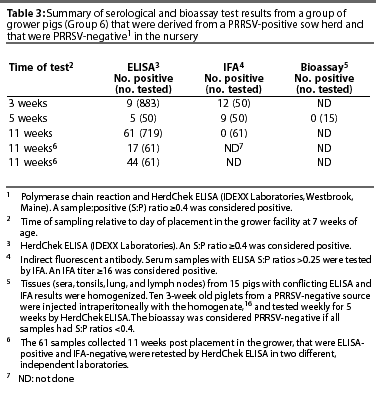
When blood was collected from the remaining 719 pigs in the group 11 weeks after placement in the grower facility, 61 animals were ELISA-positive, but all samples were IFA-negative. In addition, when the 61 ELISA-positive samples were submitted to two different laboratories for retesting using HerdChek ELISA, 17 samples tested positive in one laboratory, and 44 tested positive in the other laboratory. Because of these confusing serological results, the decision was made not to use this group of animals for stocking the new PRRSV-negative herd.
Quarantine facility
The PRRSV-negative grower groups were reassembled into five quarantine groups (Table 2), and a total of 3415 pigs were used to stock the new PRRSV-negative herd.
Three groups (1, 2B, and 7), a total of 1047 pigs, were accepted after the first ELISA test 1 week after entering the quarantine facility. All but two pigs tested negative by ELISA, and both positive samples were negative either by PCR (Group 1) or IFA (Group 7). Three weeks after they entered the quarantine facility, all pigs tested negative by ELISA, and these three groups were accepted for stocking the PRRSV-negative herd without further testing.
Seven of the total of 774 pigs in Group 3, and nine of the total of 1032 pigs in Group 4-5 (assembled from grower Groups 4A, 4B, and 5) were positive by ELISA when tested 1 week after entering the quarantine facility. All ELISA-positive animals tested negative by IFA. When retested 3 weeks later, all pigs in Group 3 were ELISA-negative. Group 3 was used for stocking the PRRSV-negative herd. In Group 4-5, four pigs tested ELISA-positive 3 weeks post entry into the quarantine facility. The four ELISA-positive samples were negative when tested by IFA. The ELISA-positive results were considered false-positive and Group 4-5 was used for stocking the PRRSV-negative herd.
Necropsy results
Necropsies were performed on a total of 500 pigs. All samples tested from sentinel pigs necropsied routinely at the end of the nursery stage were PRRSV-negative by PCR, VI, and histopathological examination. Seventy-two pigs with positive ELISA S:P ratios (suspected to be false-positives) were also submitted for necropsy, and
tissues were negative for PRRSV when tested by PCR, VI, and IHC.
False-positive ELISA tests
Across all groups selected for stocking the PRRSV-negative herd (grower and quarantine groups), 10,178 samples were tested by HerdChek ELISA. Excluding the 21 samples from the rejected groups (Groups 2A and 6), 61 ELISA results (0.59%) were considered false-positives. However, the rate of false-positives was higher for some groups. The rate of false-positives was 1.13% for grower subgroup 4B (four of 352 samples), 1.37% for grower Group 5 (17 of 1241 samples), 0.87% for Group
4-5 tested after 1 week in quarantine (nine of 1032 samples), and 7% (four of 57 samples) for Group 4-5 after 3 weeks in quarantine.
Discussion
Production of PRRSV-negative pigs from PRRSV-positive sources has been reported by several authors.3,17 In these studies, PRRSV-negative pigs were produced through isowean or SEW from sow farms known to be endemically infected with PRRSV,3 or from vaccinated, serologically stable sow herds.17 Traditional isowean or SEW procedures have proven to be inconsistent in eliminating PRRSV, as the success of these procedures depends on the dynamics of the viral infection in the sows. Therefore, isowean and SEW should be considered transitional in the establishment of PRRSV-negative farms from PRRSV-positive sources, and adequate isolation and testing of nursery and grower batches may be required.
The strategy used in this study differs from that in traditional isowean or SEW programs. In our study, PRRSV-positive gilts were first isolated from their original source in a separate location, and no additional replacement animals were added to the population. Secondly, a batching system was designed for pig flow to allow detection of PRRSV-positive batches without compromising potentially PRRSV-negative groups. In addition, extensive serial testing was performed on each batch to investigate the negative status of the pigs.
Occurrence of persistently infected animals has been described in the field and in experimentally infected animals.5,7,18 Nursery-age animals may harbor the virus in the tonsils for up to 157 days.5 Experimentally infected breeding-age animals were persistently infected, with shedding for up to 86 days post infection.7 In a field study, a very low prevalence of persistently infected sows was found following a prolonged herd-closure strategy.18 In another study, experimentally infected animals were eventually able to eliminate the viral infection after developing a protective immune response.8
In our study, the presence of actively infected gilts was relevant during the first part of the project. In order to increase the probability of producing PRRSV-negative offspring, sows were not bred until viremic animals were no longer detected by PCR. However, PRRSV was detected in three of the first six batches of pigs produced, suggesting that the virus was still active in the breeding herd. In addition, PRRSV was detected in a finishing group born in the ninth week of farrowings. No PRRSV infection was detected in later batches, suggesting that the virus had either disappeared from the breeding herd or had become inactive and non-transmissible to the offspring. Whether persistently infected gilts remained in the original donor population was not directly determined. The fact that the donor population of positive gilts was established as a discrete population of animals with no further additions probably influenced the eventual cessation of viral transmission to the offspring. Addition of replacement animals into this population might have resulted in circulation of active virus in the newly introduced animals, with production of PRRSV-positive pigs and subsequent failure of the project.
As shown by several authors, PRRSV-negative pigs are not consistently produced from PRRSV-positive gilts.3,14 In a study by Donadeu et al,14 15% of batches produced by isowean tested positive. In another study, one of eight batches also produced by isowean tested positive.3 There- fore, in our study, a segregated batching system for pig flow was used to identify positive batches without compromising the potentially negative ones. The six isolated nurseries and multiple grower facilities maximized production of negative pigs. Each batch of pigs was tested before moving to a new location. By use of this strategy, three batches of pigs were rejected at the nursery level and two more groups at the grower level, without compromising the status of the PRRSV-negative groups.
Polymerase chain reaction has been described as a useful technique to identify PRRSV-positive pigs in the early stages of infection.14 In the present study, three of the first six batches of nursery pigs were PRRSV-positive by serum PCR at either 7 or 28 days post placement in the nursery. The HerdChek ELISA test might have been substituted for the PCR technique if early detection of infected batches had not been necessary. However, identification of positive batches as early as possible was a priority in this project, justifying the use of the PCR technique.
The PCR technique failed to detect PRRSV in serum samples from a batch of nursery pigs that tested seropositive at the grower level (Group 2A). Possible explanations include slow spread of PRRSV from a positive principal to the sentinels, or lateral infection at the grower site. The source of infection for Group 2A could not be further identified.
The use of sentinels as biologic indicators of PRRSV infection was instrumental in this study. In all groups identified as PRRSV-positive, sentinels were positive either by PCR or ELISA. Testing of sentinels was preferred over testing of principal pigs at times when serological tests were expected to yield positive results because of maternal antibodies in the principal pigs. In addition, testing of the sentinels by PCR was expected to be more sensitive than testing of the principals, as maternal antibodies might have a neutralizing effect on the levels of viremia in the principal pigs,19 making identification of PRRSV-positive pigs by serum PCR potentially more difficult.
This study provided insightful information on the large-scale use of serological tests. Across all groups selected for stocking of the PRRSV-negative herd, the percentage of false-positives by the IDEXX ELISA test was 0.59%, which was within the expected range claimed by the manufacturer.13 The IDEXX ELISA test is very sensitive (100%), and specificity claimed by the manufacturer is estimated to be 99.5% (95% confidence intervals range between 98.3% and 99.9% specificity).13 However, in this study, for a given batch of pigs, the percentage of false-positives was sometimes higher than expected, and in one case was 7% (four of 57 pigs in Group 4-5 after 3 weeks in the quarantine facility). Contributing to the uneven distribution of false-positives among the groups tested might be differences in test kit batches and laboratory procedures (test were performed on different days) or another infectious agent cross-reacting with PRRSV to some degree. It is noteworthy that there were no false- positives in the 677 nursery samples tested from the accepted nursery batches. Whether the rate of false-positives increases with pig age due to nonspecific reactivity in the sera needs to be further investigated. Therefore, although the IDEXX ELISA proved to be a very good screening test on a population basis, it should not be used as the sole test for determining the PRRS status of individual animals.
In order to address the challenge presented by lack of specificity of the IDEXX ELISA test, a strategic diagnostic approach based on test, retest of the same sample, and resample of the population, was introduced at the 15th week after farrowings had started, replacing the PCR test with an alternate serological test, the IFA test, to confirm positive ELISA results. This strategy was used because it was possible that by the time an adult pig developed ELISA antibodies, it might have already cleared the viremia. Viremia in adult animals is reported to be shorter than in nursery-age pigs.20
When PCR or virus isolation is the back-up strategy for interpreting ELISA results, it is assumed that there is a correlation between the presence of antibody and virus antigen. This may be true only during the onset of infection, 10 to 35 days post infection.21 Another advantage to using the IFA as a backup for the ELISA is that both tests measure the same immunological response.15 However, the sensitivity of the IFA is low (<90%)15 and the possibility of false-negatives must be taken into account.
In our study, the low sensitivity associated with the IFA test did not appear to be a problem, as the PRRSV-negative groups remained negative in successive tests, and the newly stocked herd has remained negative for the following 18 months after stocking (unpublished data). Monthly blood samples are collected from 30 adult animals, sixty 5-month-old finishing pigs, and thirty 6-month-old breeding herd replacement animals in isolation. A total of 2020 blood samples have been tested by HerdChek ELISA, with 26 positive ELISA samples that were negative when tested by IFA. No PRRSV-positive pigs have been detected in the 18 necropsies performed, nor have clinical signs of PRRS been observed in the herd. In addition, second and third parity sows from the original positive donor population were used to stock a separate, 1800-sow farm (unpublished data). No PRRSV-positive pigs were detected either at the nursery or grower level. During the 13 months when this herd was being stocked and monitored, 811 animals were tested by HerdChek ELISA, with 16 positive samples, all of which were negative by IFA.
Serological results were inconsistent and lacked repeatability in grower Group 6, and the group was rejected because of the difficulty interpreting the serological tests, although PRRSV was not detected. The IFA- and ELISA- positive animals were PRRSV-negative by bioassay, and presence of infectious virus in the IFA- and the ELISA-positive animals could not be confirmed. However, when the remaining animals in this group were retested, a new set of 61 samples out of 719 (8.4%) were positive by ELISA, and the ELISA-positive samples were negative by IFA. In addition, when these same samples were submitted to two different laboratories for HerdChek ELISA testing, one laboratory identified 17 positive samples, and the other identified 44 positive samples. Whether this is a common situation or not needs to be further evaluated.
The obvious limitations of this study include the cost derived from the extensive testing that was conducted, and the labor and the numerous sites that were required. Retrospectively, testing could have been reduced by using a sequential strategic representative sampling instead of whole group testing (which was required for regulatory purposes). In addition, use of sows already recovered from infection as the donor population, instead of recently infected gilts, might have reduced costs, because fewer batches and pigs would have been lost due to PRRSV infection. The cost of this project was justified because PRRSV-negative elite genetics were required, but similar expense may not be justifiable for commercial swine production.
This report describes a protocol that was successful in producing PRRSV-negative pigs from PRRSV-positive sources. By creating a discrete, isolated population of positive gilts and by managing the pig flow in batches, the likelihood of producing PRRSV-negative pigs was increased and assured over time. This study also provided information on the dynamics of infection in a closed population, in which PRRSV infection, as measured by lack of seroconversion in the offspring, either disappeared or became inactive. The extensive testing conducted in this study also provided information about interpretation and large-scale implementation of diagnostic tests for PRRSV infection.
Implications
- The establishment of a PRRSV-negative population from PRRSV-positive sources was achieved by managing the positive donor gilts and batching the pig flow, allowing elite genetics in the donor herds to be preserved.
- Results of this study provide evidence that PRRSV infection, as assessed by lack of seroconversion in the offspring, may be eliminated in certain closed populations.
- The PRRSV IDEXX ELISA is a good indicator of the PRRS status of a herd, but the possibility of false-positives makes this test unreliable as the sole test determining the PRRSV status of an individual animal.
Acknowledgements
The authors would like to acknowledge Dr Marie Gramer, Dr Jose Piva, Dr Butch Baker, Dr Rick Tubbs, Dr Tom Riek, Dr Hank Harris, Dr Fernando Osorio, and Dr Jim Collins for their contributions on planning and execution of the project. The authors would also like to acknowledge all PIC personnel that participated in the project as well as the personnel at Centro de Investigacion de Salud Animal in Spain, the University of Minnesota Diagnostic Laboratory, and the Canadian Food Inspection Agency in Canada.
References - refereed
2. Wensvoort G, Terpstra C, Pol JMA, ter Lack EA, Bloemaraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Bröekhuijsen JM, Moonen PLJM, Zetstra T, de Boer EA, Tibben HJ, de Jong MF, van't Veld P, Groenland GJR, van Gennep JA, Voets MTh, Verheijde JHM, Braamskamp J. Mystery swine disease in the Netherlands: isolation of Lelystad virus. Vet Q. 1991;3:121-130.
5. Wills RW, Zimmerman JJ, Yoon K-J, Swensons SL, McGinley MJ, Hill HT, Platt KB, Christopher-Hennings, Nelson EA. Porcine reproductive and respiratory syndrome virus: A persistent infection. Vet Microbiol. 1997;55:231-240.
7. Bierk MD, Dee SA, Rossow KD, Otake S, Collins JE, Molitor TW. Transmission of porcine reproductive and respiratory syndrome virus from persistently infected sows to contact controls. Can J Vet Res. 2001;5:261-266.
10. Dee SA, Bierk MD, Deen J, Molitor TW. An evaluation of test and removal for the elimination of PRRSV from 5 swine farms. Can J Vet Res. 2001;65:22-27.
12. Alexander TJL. Medicated early weaning to obtain pigs free from pathogens endemic in the herd of origin. Vet Rec. 1980;106:114-119.
15. Yoon IJ, Joo HS, Christianson WT, Kim HS, Collins JE, Morrison RB, Dial GD. An indirect fluorescent antibody test for the detection of antibody to swine infertility and respiratory syndrome virus in swine serum. J Vet Diag Invest. 1992;4:144-147.
16. Allende R, Laegreid WW, Kutish GF, Galeota JA, Wills RW, Osorio FA. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J Virol. 2000;74:10834-10837.
18. Bierk M, Dee SA, Rossow KD, Collins JE, Guedes MI, Molitor TW. A diagnostic investigation of PRRSV persistence in adult breeding swine. Vet Rec. 2001;148:687-690.
20. Rossow KD. Porcine reproductive and respiratory syndrome. Vet Pathol. 1998;35:1-20.
References - non refereed
1. Keffaber KK. Reproductive failure of unknown etiology. AASP Newsl 1989;1:1-9.
3. Gramer ML, Christianson WT, Harris DL. Producing PRRS negative pigs from PRRS positive sows. Proc AASP. 1999;413-416.
6. Wills RW, Osorio FA, Doster AR. Transmission of PRRS virus to age-matched sentinel pigs. Proc Conf Res Workers An Diseases. 2000;141.
8. Meier WA, Wheeler J, Husmann RJ, Osorio F, Zuckermann FA. Characteristics of the immune response of pigs to wild-type PRRS virus or to commercially available vaccines: an unconventional response. Proc AASP. 2000; 415-418.
11. Torremorell M, Baker R. Eradication of PRRS virus by changing the pig flow and the introduction of negative replacements into positive sow farms. Proc A. Leman Conf. 2000;59-62.
13. HerdChek PRRS [package insert]. Westbrook, Maine: IDEXX Laboratories; 2000.
19. Osorio FA, Galeota JA, Nelson E, Brodersen B, Doster A, Wills R, Zuckermann F, Laegred WW. Passive transfer of virus-specific neutralizing antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity [abstract]. 82nd Proc Conf Res Workers An Diseases. Abstract 173.










The AASV website and The Journal of Swine Health and Production are made possible by the generous support of the AASV Industry Support Council, including: .
Copyright (C) 1996-2002 American Association of Swine Veterinarians. Please send your suggestions about this site to Dave Brown, webmaster@aasv.org.
This page last updated April 19, 2012.
