| Original research | Peer reviewed |
Cite as: Dorr PM, Gebreyes WA. Flavophospholipol: Effect on multi-drug resistant Salmonella enterica serovar Typhimurium in swine. J Swine Health Prod. 2009;17(6):308–317..
Also available as a PDF.
SummaryObjectives: To assess the effect of flavophospholipol on Salmonella shedding and antimicrobial resistance via a plasmid-curing mechanism. Materials and methods: Fifty-six pigs (60 days old) that tested negative by culture for Salmonella were allocated into four treatment groups and challenged with either Salmonella serovar Typhimurium DT104 or Salmonella Typhimurium DT193. After challenge, each treatment group was fed one of four feed rations: a ration containing no antimicrobials (Control) or the same ration containing either flavophospholipol (F), penicillin and chlortetracycline (PCt), or penicillin, chlortetracycline, and flavophospholipol (PCtF). Fecal samples were obtained weekly for 20 weeks (three samples pre-challenge and 17 post challenge) and processed for Salmonella isolation and antimicrobial-resistance testing. Results: For pigs challenged with phage type DT193, the duration of Salmonella shedding was significantly shorter in the PCtF group than in the controls, with a hazard ratio (HR) of 4.51 (P = .01). Overall, shedding time was significantly shorter in pigs challenged with DT104 than in those challenged with DT193 (HR 13.6; P < .001). The proportions of both DT104 and DT193 isolates resistant to ampicillin and amoxicillin-clavulanic acid were significantly lower after treatment with flavophospholipol. Implications: Growth-promotion levels of antimicrobials can decrease the shedding time of S Typhimurium in clinically healthy animals. Use of in-feed flavophospholipol may result in loss of resistance of S Typhimurium to certain classes of antimicrobials. | ResumenObjetivos: Valorar el efecto del flavofosfolipol en la excreción de Salmonella y resistencia antimicrobiana a través de un mecanismo de segregación de plásmido. Materiales y métodos: Se asignaron cincuenta y seis cerdos (de 60 días de edad) que resultaron negativos al cultivo de Salmonella, a cuatro grupos de tratamiento y fueron retados ya fuera con Salmonella serovariedad Typhimurium DT104 ó Salmonella Typhimurium DT193. Después del reto, se alimentó a cada grupo de tratamiento con una de cuatro raciones de alimento: una ración sin antimicrobianos (Control) ó la misma ración con flavofosfolipol (F), penicilina y clortetraciclina (PCt), ó penicilina, clortetraciclina, y flavofosfolipol (PCtF). Se obtuvieron muestras fecales semanalmente por 20 semanas (tres muestras antes del reto y 17 después del reto) y se procesaron para el aislamiento de Salmonella y resistencia antimicrobiana. Resultados: En los cerdos retados con el fago tipo DT193, la duración de la excreción de Salmonella fue significativamente más corta en el grupo PCtF que en los controles, con una tasa de riesgo (HR por sus siglas en inglés) de 4.51 (P = .01). En total, el tiempo de excreción fue significativamente más corto en cerdos retados con DT104 que en aquellos retados con DT193 (HR 13.6; P < .001). Las proporciones de ambos aislamientos DT104 y DT193 resistentes a la ampicilina y amoxicilina-ácido clavulánico fueron significativamente más bajos después del tratamiento con flavofosfolipol. Implicaciones: Niveles de promoción de crecimiento de los antimicrobianos pueden disminuir el tiempo de excreción de S Typhimurium en animales clínicamente sanos. El uso de flavofosfolipol en el alimento puede resultar en la pérdida de resistencia de la S Typhimurium contra ciertas clases de antimicrobianos. | ResuméObjectifs: Évaluer l’effet du flavophospholipol sur l’excrétion de Salmonella et sur l’antibiorésistance via un mécanisme de guérison plasmidique. Matériel et méthodes: Cinquante-six porcs (âgés de 60 jours) négatifs en culture pour la présence de Salmonella ont été répartis en quatre groupes de traitement et soumis à une infection défi soit avec Salmonella sérovar Typhimurium DT104 ou Salmonella Typhimurium DT193. Après l’infection, chaque groupe de traitement a reçu un des quatre aliments suivants: une nourriture ne contenant aucun antibiotique (Témoin) ou la même nourriture contenant soit du flavophospholipol (F), de la pénicilline et de la chlortétracycline (PCt), ou de la pénicilline, de la chlortétracycline, et du flavophospholipol (PCtF). Des échantillons de fèces ont été prélevés à chaque semaine pour une durée de 20 semaines (trois échantillons pré-infection et 17 post-infection) et analysés pour l’isolement de Salmonella et détermination de l’antibiorésistance. Résultats: Pour les porcs infectés avec le type phagique DT193, la durée de l’excrétion de Salmonella était significativement plus courte que celle des animaux témoins, avec un risque relatif (HR) de 4,51 (P = .01). De manière globale, la durée d’excrétion était significativement plus courte chez les porcs inoculés avec la souche DT104 que ceux inoculés avec la souche DT193 (HR 13,6; P < .001). Les proportions d’isolats DT104 et DT193 résistants à l’ampicilline et à l’amoxicillin-acide clavulanique étaient significativement moindres après le traitement au flavophospholipol. Implications: L’utilisation d’antimicrobiens à des concentrations de promoteur de croissance peut réduire la période d’excrétion de S Typhimurium chez des animaux cliniquement sains. L’incorporation de flavophospholipol dans les aliments peut entraîner chez S Typhimurium la perte de résistance à certaines classes d’antimicrobiens. |
Keywords: swine, Salmonella serovar Typhimurium,
antimicrobial resistance, flavophospholipol
Search the AASV web site
for pages with similar keywords.
Received: October 7, 2008
Accepted: June 29, 2009
According to the Centers for Disease Control and Prevention, Salmonella serovar Typhimurium alone was responsible for 43 major foodborne outbreaks in the United States between 1990 and 1995.1 This serovar is commonly found in swine fecal samples collected preharvest and during the harvesting process. Previous studies on Salmonella enterica, and specifically serovar Typhimurium, at swine farms and processing facilities reported its common occurrence at these stages of the production cycle.2,3 In one recent report,4 the overall prevalences of S Typhimurium on-farm and at slaughter were 57% and 27%, respectively. However, other studies have shown Salmonella prevalence to be higher at slaughter than on the farm.2,5,6
Salmonella can cause severe enteric disease in both animals and humans. Furthermore, serovar Typhimurium has been shown to be commonly multi-drug resistant (MDR).3 The DT193 and DT104 phage types used in this study are phenotypically similar; however, there is a major difference in the location of the antimicrobial resistance genes. The resistance pattern seen in the DT104 phage type is chromosomally encoded,7 while the resistance pattern seen in the DT193 is plasmid based.8 Therefore, in this study, it was important to include strains from both phage types in order to discern the effect of flavophospholipol intervention on loss of antimicrobial resistance from genes located on plasmids versus those encoded on chromosomes.
Antimicrobial resistance remains one of the leading public-health concerns. Use of antimicrobials in food animals also has been implicated as a potential factor in the dissemination of antimicrobial resistance. Withdrawal of use of antimicrobials in agriculture, particularly for growth-promotion purposes, has been recommended by some public-health organizations, and it has also been implemented in some European countries. On the other hand, it has been shown that withdrawal of antimicrobials may not have the desired effect of reversing the resistance status of bacterial strains, at least in the short term.9 Therefore, investigation of an active approach or a counter-selective agent that may be capable of selecting against antimicrobial resistance in the presence or absence of other antimicrobials is crucial. One active agent that has previously been reported to have an effect against antimicrobial resistance via plasmid-curing mechanism is flavophospholipol.10 This antimicrobial agent is approved for use in feed for swine. Furthermore, there is currently no compound being used in human medicine that is an analog of flavophospholipol, and although its mechanism of action is similar to that of the β-lactam family, no cross-resistance has been reported.
A recent study involving pathogens, including Escherichia coli and Enterococcus species, established that flavophospholipol has a plasmid-curing effect that resulted in a reduction of antimicrobial-resistant E coli.10 In 1976, Dealy and Moeller11 showed that use of flavophospholipol resulted in decreased Salmonella shedding in pigs, as well as decreased resistance to different classes of antimicrobials.11,12 However, there is a paucity of studies that have replicated these initial observations, particularly with contemporary MDR Salmonella strains that exhibit an array of multi-drug resistance profiles. In this study, we evaluated two MDR S Typhimurium strains with pandemic distribution: phage type DT104 resistance-type (R-type) ACSSuTAx (resistant to ampicillin [A], chloramphenicol [C], streptomycin [S], sulfisoxizole [Su], tetracycline [T], and amoxicillin-clavulanic acid [Ax]) and phage type DT193 R-type AKSSuT (resistant to ampicillin, kanamycin [K], streptomycin, sulfisoxizole, and tetracycline).3
The specific aims of this study were to determine the effect of flavophospholipol in vivo on the prevalences and phenotypic profiles of two predominant MDR Salmonella strains (S Typhimurium DT104 and DT193) in pigs exposed to flavophospholipol and other antimicrobials in the feed. We also compared and assessed differences in the counter-selective action of flavophospholipol on a plasmid-borne MDR strain (S Typhimurium DT193) and a chromosomally encoded MDR strain (S Typhimurium DT104).8
Materials and methods
Approval was received from the Institutional Animal Care and Use Committee of North Carolina State University, Raleigh, North Carolina, to conduct the study as per the described protocol and procedures.
Animals
Pigs that had not been exposed to antimicrobials since birth, originating from the North Carolina State University institutional farm unit, were chosen for this study at approximately 3 weeks of age. A total of 56 pigs tested negative for Salmonella three times using serial culturing on fecal samples and an enrichment method on Days -32, -30, and -16 of the study (Figure 1).
Figure 1: Timeline for study in which pigs inoculated with Salmonella serovar Typhimurium were treated with in-feed antimicrobials or were untreated controls.
Ct = chlortetracycline; F = flavophospholipol; P = penicillin |
Challenge group preparation
Salmonella Typhimurium strains DT104 (isolate #UF19) and DT193 (isolate #UT8) were plated onto Luria Bertani (LB) agar (Becton, Dickinson and Company, Sparks, Maryland) from frozen stock and incubated at 37°C for 24 hours. A colony of each isolate was then selected and transferred to 25 mL of LB broth with 25 μg per mL tetracycline and incubated for 24 hours in a shaker bath at 37°C. Challenge concentration was then adjusted to 1 × 108 colony forming units (CFU) per mL. At the experimental site, 5-mL aliquots of challenge material were transferred to 10-mL syringes, which were used to inoculate the pigs orally. The DT193 challenge group received an average dose of 8.14 × 108 bacteria and the DT104 group received an average dose of 2.55 × 108 bacteria, consistent with challenge doses in previous studies.13,14
Nursery-aged pigs (n = 56) were randomly assigned to their respective treatment groups using a random number generator (random.org, available at www.random.org). All pigs received injections of ceftiofur (Excede; Pfizer, New York, New York; 50 mg) 35 and 23 days prior to in-feed antibiotic intervention (Figure 1) to aid in clearing Salmonella that may have been present but not detected, to disrupt enteric flora, and to facilitate colonization of the challenge strains of Salmonella. The 56 pigs were allocated into two challenge groups, each challenged with one of the two MDR strains of S Typhimurium: DT104 (R-type ACSSuTAx) or DT193 (R-type AKSSuT). Each isolate was tested using the Kirby-Bauer method15,16 prior to challenge to ensure phenotype continuity.
Study design
Pigs were allocated into eight treatment groups: DT104 challenge with four antimicrobial interventions (Control, F, PCt, and PCtF) and DT193 challenge with the same four antimicrobial interventions. From weaning (approximately 21 days of age) until 2 weeks before challenge (approximately 49 days of age), the pigs were maintained in a conventional nursery facility with rubber-coated, slotted-steel floors and were fed a ration free of antimicrobials.
At the challenge facility, animals were housed in two separate but identical rooms, each containing eight pens, with three or four pigs per pen. Each pen had a solid slanted concrete floor with concrete sidewalls, preventing nose-to-nose contact between pigs in neighboring pens. Pigs were housed in rooms by challenge strain (DT193 or DT104) and were penned by treatment group. Each pig was challenged with one of the Salmonella phage types on Day -8 of the study. Fecal samples were collected from each pig three times (Days -7, -4, and -2) before antimicrobial intervention began on Day 0, then once a week in the post-intervention period until the animals were humanely euthanized approximately 15 to 17 weeks later. Strain DT104 appeared not to colonize well (ie, repeated fecal cultures from multiple pigs were negative), so the pigs challenged with this strain were challenged a second time on Day 0 of the study, prior to antimicrobial intervention.
From Day 0 until week 15 to 17 of the study (when pigs were approximately 24 to 26 weeks of age), the study subjects were fed a formulated finishing diet with interventions as shown in Table 1. Group C received the base diet, to which no antimicrobials were added. For the three treated groups, antimicrobials were added to the control ration, including flavophospholipol (Flavomycin; Huvepharma Inc, Peachtree City, Georgia) at 4.85 g per tonne (Group F); penicillin and chlortetracycline, both at 11 g per tonne (Group PCt); and penicillin and chlortetracycline, both at 11 g per tonne, plus flavophospholipol at 4.85 g per tonne (Group PCtF).
Table 1: Challenge strains and interventions in a study in which 56 pigs inoculated with Salmonella serovar Typhimurium (Day -8) were treated with in-feed antimicrobials for 15-17 weeks, beginning Day 0, or were untreated controls
* Dosages: flavophospholipol, 4.85 g/tonne; penicillin, 11 g/tonne; chlortetracycline, 11 g/tonne. † Resistance- (R-) type abbreviations: A = ampicillin; C = chloramphenicol; S = streptomycin; Su = sulfasoxazole; T = tetracycline; Ax = amoxicillin-clavulanic acid; K = kanamycin. |
|||||||||||||||||||||||||||||||||
The investigators were blinded to treatment until the conclusion of the study. Strict biosecurity measures for personnel attending the pigs and facilities were enforced during the study period.
Sample collection and processing
Fecal samples from pigs were transported to our laboratory (College of Veterinary Medicine, North Carolina State University), processed, and cultured. All samples were collected in sterile plastic cups under conditions minimizing sample and animal cross-contamination. Clean disposable coveralls and disposable boots were worn into the rooms, and personnel blinded to treatment were assigned to either the DT104 pigs or the DT193 pigs to prevent cross-contamination.
Upon arrival at the abattoir, the animals were rested on the trailer to avoid exposure to cross-contaminants in the holding pens. Cecal contents (10 g) and mesenteric lymph-node samples (10 g) were collected from each carcass. Instruments used to collect these materials were sterilized by flaming with ethanol between samples.
Salmonella isolation
All samples were processed for Salmonella isolation as described previously.17 A maximum of five Salmonella colonies were selected from each of the presumptive positive xylose lysine tergitol 4 (XLT4) plates and were subjected to further biochemical testing for triple sugar iron (TSI) and urea for confirmation. After the isolates were confirmed to be Salmonella, antimicrobial susceptibility testing was performed on each isolate.
Antimicrobial susceptibility testing
Isolates were tested using the Kirby-Bauer disc diffusion method as recommended by the Clinical Laboratory Standards Institute (CLSI)15,16 and as described previously.8 The following antimicrobials and disc potencies were used: ampicillin (10 μg), amoxicillin-clavulanic acid (30 μg), amikacin (30 μg), ceftriaxone (30 μg), cephalothin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), kanamycin (30 μg), streptomycin (10 μg), sulfisoxazole (250 μg), and tetracycline (30 μg). Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, and Pseudomonas aeroginosa ATCC 27853 were used as quality-control organisms.
Genotyping by pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) analysis was performed on selected isolates to confirm the clonality between the initial challenge strains and those recovered during the study (Figure 2). The protocol used was as recommended previously by the PulseNet, CDC (http://www.cdc.gov/pulsenet/). Briefly, 200 mL overnight culture cells from tryptic soy agar were lysed, and intact genomic DNA was digested in agarose-embedded plugs with Xba1 restriction enzyme. The digested DNA was then separated using a contour-clamped homogeneous electric field (CHEF)-DRIII (Bio-Rad Laboratories, Hercules, California) with the following conditions: 0.5× TBE (Tris-borate EDTA), 1% Seakem Gold agarose (Seakem, FMC Corporation, Philadelphia, Pennsylvania), 14°C, and 6V per cm for 18 hours with switch times ranging from 2.2 to 63 seconds. Gels were stained with ethidium bromide for 30 minutes, destained three times for 20 minutes each with distilled water, and photographed using Alpha Imager (Alpha Innotech Corporation, San Leandro, California). Analysis of PFGE data was performed using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) using the “different bands” algorithm for clustering and the unweighted pair group for arithmetic means tree-building approach with optimization of 1% and 0.75% position tolerance. Visual inspection of the patterns was performed as a final step for analysis.
Figure 2: Pulsed-field gel electrophoresis dendrogram of Salmonella DT193 isolates obtained from fecal-culture samples during a study in which pigs were challenged with strain DT193 (Day -8; approximately 64 days of age) and treated with flavophospholipol for 15 to 17 weeks beginning on Day 0, as described in Figure 1 and Table 1 This figure shows phenotypic diversity but genotypic clonality with the original challenge strain (identified as “Pig #0”). A single contaminant strain (ID 6490) was detected from one pig in the Control group. Under “Treatment,” C = Control group, F = flavophospholipol group; under “R-type,” A = ampicillin, S = streptomycin, Su = sulfasoxazole, T = tetracycline, and K = kanamycin. 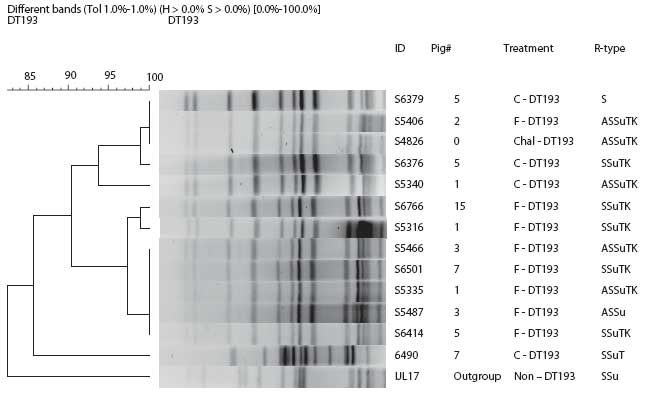 |
Statistical analysis
The proc freq (chisq cmh nocol nopct) method in the SAS software package (SAS Institute, Inc, Cary, North Carolina) was used to estimate the differences in percentages of resistance among treatment groups and changes between groups and between phage types, using the chi-square statistic. To assess the degree of deviation from control among different phenotypes, odds ratios and 95% confidence intervals were constructed using this formula:
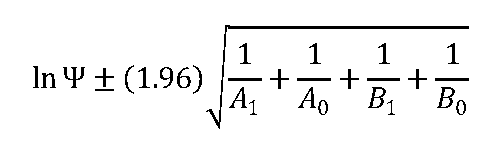
To assess the difference in prevalence through time, Cox proportional hazards survival analysis was used in the EGRET software package (Cytel Inc, Cambridge, Massachusetts) to obtain the survival graphs and hazard ratios (HRs). Values of P < .05 were considered statistically significant. Note that animals were defined as “not shedding” when they tested negative for Salmonella, using the isolation method described, for two consecutive fecal collections.
Results
Salmonella shedding times for the two challenge strains
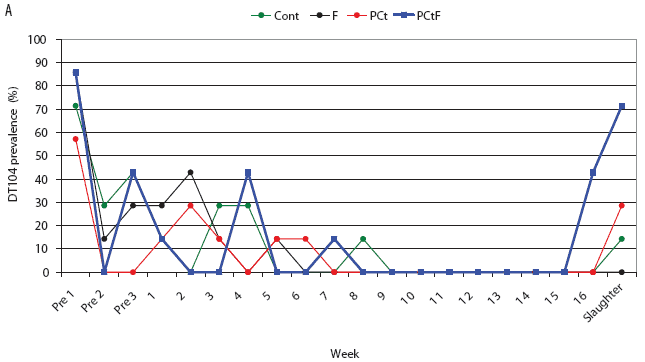
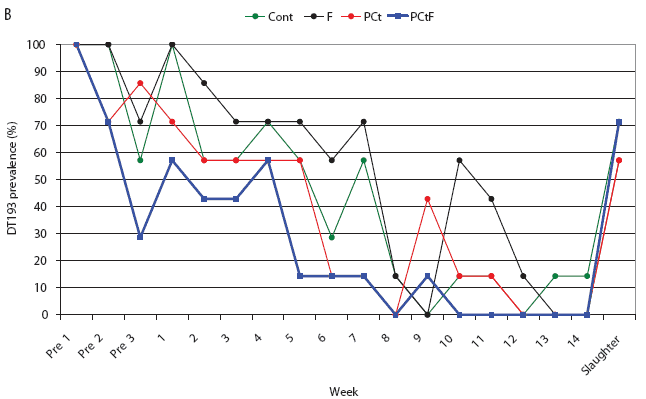
For the treatment groups challenged with phage type DT193, only the PCtF group had a significantly shorter duration of Salmonella shedding than the control group, with an HR of 4.51 (P = .01). Median survival times (ie, days of Salmonella shedding) were estimated to be 24, 45, 59, and 59 days for the PCtF, PCt, F, and Control groups, respectively. In both the DT193 and DT104 challenge groups, shedding times for the F and PCt treatments did not differ significantly from those of the Control groups. Overall, pigs challenged with the DT104 strain had a significantly shorter shedding time than did pigs challenged with the DT193 strain (HR 13.6; P < .001), with approximate median survival times of 4 and 43 days respectively. Prevalence of pigs with positive fecal samples over the course of the study is depicted in Figure 3.
Figure 3: Salmonella-positive fecal culture prevalence by week for the study described in Figure 1 and Table 1. Pigs were challenged at approximately 64 days of age (Pre 1) with Salmonella serovar Typhimurium phage type DT104 (A) or DT193 (B). Antimicrobial intervention began after Pre 3. Cont = control, no antimicrobial in feed; F = flavophospholipol in feed at 4.85 g per tonne; PCt = penicillin and chlortetracycline, both in feed at 11 g per tonne; PCtF = penicillin and chlortetracycline, both in feed at 11 g per tonne, plus flavophospholipol at 4.85 g per tonne.
|
Loss of resistance to individual antimicrobials
For the treatment groups challenged with the Salmonella DT193 phage type, there was no significant difference in loss of resistance to ampicillin among groups. However, when considering all isolates in all groups, there was an overall loss of resistance in 54 of 1021 isolates (5.3%). For the treatment groups challenged with the DT104 phage type, loss of ampicillin resistance was significantly greater in the PCt group than in the F group (P < .001) and Control group (P = .03). Significantly fewer ampicillin-resistant isolates were recovered from the PCtF group than from the F group (P = .001). Also, when groups with the same intervention were compared for different challenge strains, there was a significantly greater loss of ampicillin resistance in the PCt and PCtF treatments in the DT104 challenge group than in the DT193 group (Table 2).
Table 2: Antimicrobial resistance in two Salmonella serovar Typhimurium phage types recovered from fecal samples from groups of pigs (n = 7) inoculated with Salmonella serovar Typhimurium on Day -8 and treated with in-feed antibiotics for 15-17 weeks, beginning Day 0, or not treated*
* Treatment groups described in Figure 1 and Table 1. † At the beginning of the study, 100% of both challenge strains were resistant to the four listed antimicrobials. No. of tests varies because of antimicrobial discs lost during culturing. ‡ Chi-square analysis. F = Flavophospholipol; PCt = Penicillin, chlortetracycline; PCtF = Penicillin, chlortetracycline, flavophospholipol. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
For the DT193 phage type, streptomycin and sulfisoxazole resistance were maintained throughout the study in 100% of the isolates recovered from all treatment groups. In the DT104 challenge group, frequency of resistance to these antimicrobials was significantly lower in the isolates recovered from the Control group than from either the F or PCtF group (P < .05).
For the DT193 phage type, significantly fewer tetracycline-resistant isolates were recovered from the Control and PCtF groups than from the F group (P = .01 in each case). Also, there was a trend (P < .10) toward fewer tetracycline-resistant isolates in the PCt group than in the F group. For the treatment groups challenged with the DT104 phage type, there was no significant difference in loss of resistance to tetracycline among groups. However, there was an overall loss of resistance to tetracycline among all groups, with an average of 10% of isolates (29 of 296) recovered in each group having been rendered susceptible to tetracycline. Furthermore, significantly fewer tetracycline resistant isolates were recovered from the DT104 F and PCt groups than from the DT193 F (P = .003) and PCt groups (P = .01).
Resistance of phage type DT104 to amoxicillin-clavulanic acid was significantly lower in the isolates obtained from the F treatment group than in isolates obtained from either the Control (P < .001) or PCtF treatments (P = .049), and was significantly lower in the isolates from the PCt group than in those from the Control group (P < .01).
Analysis of loss of MDR profiles
When the isolates were analyzed on the basis of phenotype, significantly more DT193 isolates from the F group showed a shift in antimicrobial resistance pattern (R-type) to KSSuT (from the challenge AKSSuT) than did the Control group (OR 3.64; 95% CI, 1.03-12.84; Figure 4). In addition, there were significantly fewer isolates with “S” phenotypes (ie, resistant to streptomycin alone) in the F group than in the Control group (OR 0.051; 95% CI, 0.002-0.915). Among the DT104 challenge pigs, significantly more isolates were recovered that were phenotypically identical to the challenge strain in the Control group than in any other group. Only in the F group were significantly more isolates recovered that lost resistance specifically to amoxicillin-clavulanic acid than in the Control (OR 7.5; 95% CI, 1.6-36.1; Figure 5).
Figure 4: Post antimicrobial intervention phenotypic analysis of Salmonella isolates recovered from treatment groups challenged with Salmonella serovar Typhimurium DT193 as described in Figure 1 and Table 1. A = ampicillin, C = chloramphenicol, S = streptomycin, Su = sulfisoxazole, T = tetracycline, K = kanamycin. Cont = Control, no antimicrobial in feed; F = flavophospholipol in feed at 4.85 g per tonne; PCt = penicillin and chlortetracycline, both in feed at 11 g per tonne; PCtF = penicillin and chlortetracycline, both in feed at 11 g per tonne, plus flavophospholipol at 4.85 g per tonne. Indicated proportions (*) were significantly different from the Control (P < .05; chi-square). 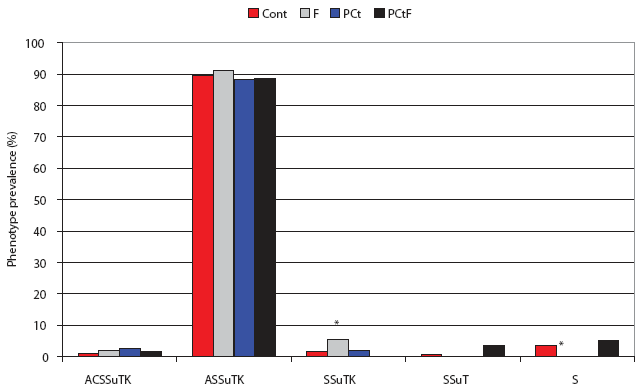 |
Figure 5: Post antimicrobial intervention phenotypic analysis of Salmonella recovered from treatment groups challenged with Salmonella Typhimurium DT104 as described in Figure 1 and Table 1. A = ampicillin, C = chloramphenicol, S = streptomycin, Su = sulfisoxazole, T = tetracycline, Ax = amoxicillin-clavulanic acid, and Cf = cephalothin. Cont = Control, no antimicrobial in feed; F = flavophospholipol in feed at 4.85 g per tonne; PCt = penicillin and chlortetracycline, both in feed at 11 g per tonne; PCtF = penicillin and chlortetracycline, both in feed at 11 g per tonne, plus flavophospholipol at 4.85 g per tonne. Indicated proportions (*) were significantly different from the Control (P < .05; chi-square). 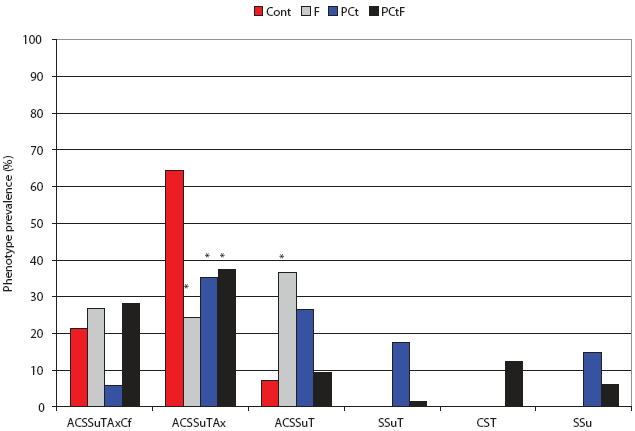 |
To confirm the clonality of the DT193 challenge strain with isolates that showed change in antimicrobial resistance profile, DNA fingerprinting was performed using PFGE on four selected isolates from the control group and eight selected isolates from the F group. Results confirm that the isolates were predominantly clonal (Figure 5). One isolate from the Control group was a contaminant not related to the challenge strain. This finding confirms that the change in antimicrobial resistance profile of the F group occurred predominantly in the challenge strain itself and was not caused by introduction of a new strain. In the DT104 challenge groups, those that gained cephalosporin resistance were clonal with the challenge strain (data not shown).
Discussion
In this study, we tested the hypotheses that feeding flavophospholipol to pigs decreases overall duration of Salmonella shedding and has an plasmid-curing effect that alters the phenotypic profile of select MDR S Typhimurium strains. Previous studies have shown that flavophospholipol has the ability to inhibit the transfer of plasmids containing antimicrobial-resistance genes.18 A study by Dealy and Moeller11 showed a significant decrease in the number of Salmonella isolates obtained from pigs that were fed flavophospholipol over a period of time. Also, in this same study,11 the Salmonella isolates recovered from the flavophospholipol group had significantly less resistance to ampicillin, streptomycin, triple sulfa, and oxytetracycline. Furthermore, there is speculation that flavophospholipol has a plasmid-curing effect on bacteria that contain resistance genes on plasmids; however, the exact mechanism has never been proven.19 The results from these previous studies,11,18,19 combined with the fact that the flavophospholipol compound has no human or animal therapeutic counterpart, makes it a favorable choice for a feed additive when intervention is needed for the highly prevalent MDR S Typhimurium.
The findings in this study indicate that, regardless of the challenge strain, the use of antibiotics resulted in a shorter shedding duration. The group that was exposed to all three antimicrobials (PCtF group) had a significantly shorter duration of shedding than the Control group. This correlation may be due to a synergistic effect among the antimicrobials, as suggested previously.20,21 All three antimicrobials were used at growth-promotion levels. This finding reaffirms the fact that the use of antibiotics may have an effect on the survival and duration of bacterial shedding when fed at levels recommended strictly for growth promotion.22 The findings also indicated that the duration of shedding varies between the two strains. The DT193 phage type was shed longer than the DT104, even though both phage types were present when the pigs were euthanized.
When the difference in loss of resistance was analyzed for individual antimicrobials, the overall loss of resistance between treatment groups and the direct effect on the phenotype was hard to appreciate. It is with the phenotypic analysis between the groups that we noticed a significant loss of resistance, specifically to ampicillin8 and amoxicillin-clavulanic acid among the isolates in the F treatment. This finding is made stronger by the fact that these isolates were proven to be clonal by the use of PFGE. The F group was the only group that had isolates with a significant phenotypic deviation when compared to the Control group.
Some isolates gained resistance to cephalothin. The appearance of the cephalosporin resistance in some of the isolates in the DT104 group may be due to an over-expression of the blaPSE1 gene, which encodes for resistance against ampicillin and possibly amoxicillin-clavulanic acid in the β-lactam group of antibiotics,7-9 or it may be an effect of decreased OmpF, OmpD, and OmpC porin permeability in the bacterial membrane, as described previously.20,21
The loss of ampicillin resistance was greater in the PCt group than in the F and Control groups within the DT104 challenge category. This finding is contrary to our conventional understanding that the selective pressure imposed by penicillin use results in maintenance of ampicillin resistance. In contrast, the findings showed that the Control groups maintained ampicillin resistance. As DT104 encodes its resistance chromosomally, it is expected that the resistance phenotype is often maintained regardless of antimicrobial use. However, it is not clear how and why some of the isolates from the PCt group showed variations in the resistance phenotype.
The loss of ampicillin and streptomycin resistance was greater in the DT104 challenge group than in the DT193 group. The DT193 isolates that were exposed to the F treatment showed loss in their pentaresistance architecture. The most common change was from AKSSuT to KSSuT. This change could be due to the hypothesized plasmid-curing effect. The DT193 strain used in this study previously has been shown to have more than one plasmid carrying various antimicrobial-resistance genes, including one with ampicillin resistance.8
Overall, flavophospholipol alone did not significantly decrease the shedding time of the challenge strains of bacteria. However, it did have a significant effect on both S Typhimurium phage types DT193 and DT104 by specifically reducing the proportion of isolates that were resistant to ampicillin and amoxicillin-clavulanic acid, respectively. Ideally, gnotobiotic pigs would have been used in this study, and there is some concern that the contamination found in the Control group may have skewed some aspects of the findings towards the null hypothesis.
Implications
- Growth-promoting levels of penicillin, chlortetracycline, and flavophospholipol can decrease the shedding time of S Typhimurium in clinically healthy animals.
- Treatment with in-feed flavophospholipol may result in the loss of resistance (change in phenotype or antibiogram) of S Typhimurium to certain classes of antimicrobials.
References
1. CDC - Foodborne Outbreak Response and Surveillance Unit. US foodborne disease outbreaks. 2002;2006:1. Available at: http://www.cdc.gov/foodborneoutbreaks/outbreak_data.htm. Accessed 15 September 2009.
2. Dorr PM, Tadesse DA, Zewde BM, Thakur S, Gebreyes WA. Longitudinal study on Salmonella dispersion and the role of environmental contamination in commercial swine production systems. Appl Environ Microbiol. 2009;75:1478–1486.
3. Gebreyes WA, Thakur S, Davies PR, Funk JA, Altier C. Trends in antimicrobial resistance, phage types and integrons among Salmonella serotypes from pigs, 1997–2000. J Antimicrob Chemother. 2004;53:997–1003.
*4. Rostagno MH, McKean JD. Prevalence of Salmonella enterica and Salmonella enterica serotype typhimurium in swine at slaughter. Proc Res Work Anim Dis Conf. 2006;439–440.
5. Hurd HS, McKean JD, Griffith RW, Wesley IV, Rostagno MH. Salmonella enterica infections in market swine with and without transport and holding. Appl Environ Microbiol. 2002;68:2376–2381.
6. Larsen ST, McKean JD, Hurd HS, Rostagno MH, Griffith RW, Wesley IV. Impact of commercial preharvest transportation and holding on the prevalence of Salmonella enterica in cull sows. J Food Prot. 2003;66:1134–1138.
7. Briggs CE, Fratamico PM. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849.
8. Gebreyes WA, Altier C. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J Clin Microbiol. 2002;40:2813–2822.
9. Kachatourians GG. Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria. Can Med Assoc J. 1998;159:1129–1136.
10. van den Bogaard AE, Hazen M, Hoyer M, Oostenbach P, Stobberingh E. Effects of flavophospholipol on resistance in fecal Escherichia coli and enterococci of fattening pigs. Antimicrob Agents Chemother. 2002;46:110–118.
11. Dealy J, Moeller MW. Influence of bambermycins on Salmonella infection and antibiotic resistance in swine. J Anim Sci. 1976;42:1331–1336.
12. Pfaller MA. Flavophospholipol use in animals: Positive implications for antimicrobial resistance based on its microbiologic properties. Diagn Microbiol Infect Dis. 2006;56:115–121.
13. Loynachan AT, Harris DL. Dose determination for acute Salmonella infection in pigs. Appl Environ Microbiol. 2005;71:2753–2755.
14. Hurd HS, Gailey JK, McKean JD, Rostagno MH. Rapid infection in market-weight swine following exposure to a Salmonella typhimurium-contaminated environment. Am J Vet Res. 2001;62:1194–1197.
15. National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disc and dilution susceptibility tests for bacteria isolated from animals; approved standard. 2nd ed. M31-A2. Wayne, Pennsylvania: National Committee for Clinical Laboratory Standards; 2002.
16. National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. 12th informational supplement, M100-S12. Wayne, Pennsylvania: National Committee for Clinical Laboratory Standards; 2002.
17. Gebreyes WA, Davies PR, Morrow WE, Funk JA, Altier C. Antimicrobial resistance of Salmonella isolates from swine. J Clin Microbiol. 2000;38:4633–4636.
18. Butaye P, Devriese LA, Haesebrouck F. Influence of different medium components on the in vitro activity of the growth-promoting antibiotic flavomycin against enterococci. J Antimicrob Chemother. 2000;46:713–716.
19. Jukes TH, DuPont HL, Crawford LM, eds. Antibiotics, Sulfonamides, and Public Health. Boca Raton, Florida: CRC Press, Inc; 1984.
20. Bellido F, Vladoianu IR, Auckenthaler R, Suter S, Wacker P, Then RL, Pechere JC. Permeability and penicillin-binding protein alterations in Salmonella muenchen: Stepwise resistance acquired during beta-lactam therapy. Antimicrob Agents Chemother. 1989;33:1113–1115.
21. Medeiros AA, O’Brien TF, Rosenberg EY, Nikaido H. Loss of OmpC porin in a strain of Salmonella typhimurium causes increased resistance to cephalosporins during therapy. J Infect Dis. 1987;156:751–757.
22. Girard AE, English AR, Evangelisti DG, Lynch JE, Solomons IA. Influence of subtherapeutic levels of a combination of neomycin and oxytetracycline on Salmonella typhimurium in swine, calves, and chickens. Antimicrob Agents Chemother. 1976;10:89–95.
* Non-refereed reference.