| Original research | Peer reviewed |
SummaryObjective: To assess genetic and antigenic properties of contemporary human-like H1 (δ-cluster) swine influenza virus (SIV) isolates circulating in US swine herds. Materials and methods: The hemagglutinin genes from 37 δ-cluster SIV isolates were fully sequenced. The isolates were antigenically categorized using a high throughput serum neutralization (HTSN) assay incorporating antisera generated against the four currently circulating clusters of H1 SIV and a commercial vaccine containing a δ-cluster SIV isolate. A subset of the isolates were further characterized with the hemagglutination inhibition (HI) assay using H1 SIV antisera. Results: Genetic analysis of the hemagglutinin gene of 2009-2010 δ-cluster isolates identified five distinct subclusters with 97% to 100% sequence similarity within subclusters and 95% to 97% similarity between subclusters. Antisera generated against SIV representing α, β, and γ clusters failed to neutralize any of 37 δ-cluster viruses in an HTSN assay. Only 46% of the δ-cluster isolates were neutralized by at least one δ-cluster antiserum. Hemagglutination inhibition assay results on a subset of thirteen 2010 isolates were in good agreement with the HTSN assay, with 38% of the isolates positive (HI titer > 40) to at least one δ-cluster antiserum. There was no measurable antibody titer to antiserum generated from the commercial vaccine or the 13 isolates. Implications: Significant genetic and antigenic heterogeneity exists among δ-cluster H1 SIV isolates, suggesting that either multiple representatives of this cluster may be required in commercial vaccines or that herd-specific vaccines may be required to protect swine from influenza virus infection. | ResumenObjetivo: Valorar las propiedades genéticas y antígenas de los aislados del virus contemporáneo de la influenza porcina H1 (grupo-δ) (SIV por sus siglas en inglés) humano-similar circulando en los hatos porcinos de EUA. Materiales y métodos: Se secuenciaron los genes de la hemaglutinina de 37 aislados de la SIV de grupo-δ. Los aislados se categorizaron antigénicamente utilizando una prueba de seroneutralización de alto rendimiento (HTSN por sus siglas en inglés) incorporando antisuero generado contra las cuatro grupos de SIV H1 que circulan actualmente y una vacuna comercial que contiene un aislado de SIV grupo-δ. Después se caracterizó un subconjunto de los aislados con la prueba de inhibición de hemaglutinación (HI por sus siglas en inglés) utilizando antisuero SIV H1. Resultados: Los análisis genéticos del gen de hemaglutinina de los aislados de grupo-δ 2009-2010 identificaron cinco subgrupos distintos con 97% a 100% de similitud de secuencia dentro de subgrupos y 95% a 97% de similitud entre subgrupos. El antisuero generado contra la SIV representando los grupos α, β, y γ falló en neutralizar cualquiera de los 37 virus de grupo-δ en una prueba de HTSN. Sólo el 46% de los aislados de grupo-δ se neutralizaron con por lo menos un antisuero de grupo-δ. Los resultados de la prueba de inhibición de la hemaglutinación en un subgrupo de trece aislados 2010 concordaron con la prueba de HTSN, con 38% de los aislados positivos (título HI > 40) a por lo menos un antisuero grupo-δ. No hubo títulos de anticuerpos medibles al antisuero generado de la vacuna comercial ó de los 13 aislados. Implicaciones: Existe heterogeneidad genética y antígena significativa entre los aislados SIV H1 de grupo-δ, sugiriendo que pueden requerirse múltiples representantes de este grupo en vacunas comerciales ó que pueden requerirse vacunas específicas para hato para proteger a los cerdos contra la infección del virus de la influenza. | ResuméObjectif: Évaluer les propriétés génétiques et antigéniques d’isolats contemporains du virus de l’influenza porcin (SIV) apparentés au type humain H1 (agrégat-δ) circulant dans les troupeaux porcins aux États-Unis. Matériels et méthodes: Les gènes de l’hémagglutinine de 37 isolats de SIV agrégat-δ ont été complètement séquencés. La catégorisation antigénique des isolats a été effectuée au moyen d’une épreuve de séro-neutralisation à débit élevé (HTSN) incorporant des antisérums dirigés contre les quatre agrégats de SIV H1 circulant actuellement et un vaccin commercial contenant un isolat de l’agrégat-δ de SIV. Un groupe accessoire de ces isolats a été caractérisé plus à fond par une épreuve d’inhibition de l’hémagglutination (HI) à l’aide d’antisérums dirigés contre H1 du SIV. Résultats: L’analyse génétique du gène de l’hémagglutinine des isolats 2009-2010 de l’agrégat-δ a permis d’identifier cinq sous-agrégats distincts ayant 97% à 100% de similarité de séquence à l’intérieur des sous-agrégats et 95% à 97% de similarité entre les sous-agrégats. Les antisérums générés contre les SIV représentant les agrégats α, β, et γ n’ont neutralisé aucun des 37 virus de l’agrégat-δ dans l’épreuve HTSN. Seulement 46% des isolats de l’agrégat-δ étaient neutralisés par au moins un des antisérums contre l’agrégat-δ. Les résultats de l’HI sur un sous-groupe de 13 isolats de 2010 étaient en accord avec les résultats de l’épreuve HTSN, avec 38% des isolats positifs (titre HI > 40) à au moins un des antisérums contre l’agrégat-δ. Il n’y avait aucun titre d’anticorps mesurables générés par le vaccin commercial ou les 13 isolats. Implications: Une hétérogénéité génétique et antigénique significative existe parmi les isolats de SIV H1 agrégat-δ, ce qui suggère que de multiples représentants de cet agrégat pourraient être requis pour inclusion dans les vaccins commerciaux ou que des vaccins spécifiques au troupeau pourraient être requis pour protéger les porcs d’une infection par le virus de l’influenza. |
Keywords: swine, swine influenza virus, serum neutralization, hemagglutination inhibition, SIV
Search the AASV web site
for pages with similar keywords.
Received: August 9, 2010
Accepted: February 28, 2011
Swine influenza virus (SIV) has shown a propensity for reassortment. While classic H1N1 SIV was the dominant subtype of influenza present in pigs in the 20th century, the introduction of H3N2 into swine herds in 1998 marked the beginning of increasing SIV genetic diversity.1-3 The H3N2 viruses were subsequently shown to be triple reassortants composed of genes from human, avian, and swine influenza virus origin.4 This particular constellation of internal genes, designated triple reassortant internal gene (TRIG), has proven particularly advantageous to SIV.5 Reassortment of the hemagglutinin (HA) gene from classic H1N1 and H3N2 led to the discovery of triple reassortant H1N2 SIV in Indiana in 1999.6,7 Another reassortment with the HA and neuraminidase (NA) genes from classic H1N1 and triple reassortant H3N2 resulted in triple reassortant H1N1 SIV.2 Triple reassortant H1N1 and H1N2 SIV have since become endemic in US swine herds.8 Additionally, H3N1 SIV, formed by reassortment between H3N2 and classic H1N1, was identified in 2004, but has not become widespread in swine herds.9
Continued introductions of new viral genetic material into the swine herd have been documented. In an isolated incident in 2006, an avian-swine H2N3 reassortant was isolated from a farm in Missouri.10 More recently, the pandemic H1N1 of 2009 resulted from a triple reassortant H1 SIV that acquired Eurasian lineage NA and matrix (M) genes.11
In 2003, wholly human and human-swine double reassortants were identified in Canada.12 Soon thereafter, H1N1 and H1N2 TRIG SIV were identified with human lineage HA and NA. These “human-like” SIVs (δ-cluster) have become endemic in swine herds. Genetic analyses of SIV isolates in the authors’ laboratory indicates that 31% of H1 SIVs isolated from April 2009 to March 2010 contain human lineage HA. Genetic and antigenic analysis of four δ-cluster SIV isolates found that this cluster is antigenically distinct from triple reassortant H1N1 and H1N2 SIVs bearing swine lineage HA.13
A high throughput serum neutralization (HTSN) assay to antigenically categorize SIVs was recently described for H3N2 SIVs.14 This methodology makes use of reference antisera to antigenically categorize SIVs in a modified serum neutralization assay. Incorporation of antisera generated against multiple isolates of circulating SIV subtypes allows assessment of antigenic diversity. Routine HTSN analysis of SIV isolates found that a substantial number of δ-cluster H1 SIVs were not neutralized by any of the three δ-cluster H1 SIV antisera routinely used in our laboratory, suggesting considerable antigenic diversity exists within this cluster. The objective of this study was to genetically and antigenically characterize δ-cluster SIVs isolated over a 12-month period to ultimately improve control measures against this SIV subtype.
Materials and methods
All animal care and handling and the experimental protocol were approved by Newport Laboratories Institutional Animal Care and Use Committee.
Viruses
All viruses were field strains isolated from clinical samples (nasal swabs or lung tissue) submitted to Newport Laboratories for isolation and genetic analysis in 2009 and 2010 as part of routine diagnostic testing. It is unknown whether samples originated from nonvaccinated animals or represented vaccine failures. Samples originated from North Carolina, Indiana, Ohio, Illinois, Minnesota, Iowa, Oklahoma, and Texas (Figure 1). Standard reference SIV sequences were used to identify the four clusters of H1 SIV: classic H1N1 (α cluster), A/swine/Indiana/88; H1N2-like (γ cluster), A/swine/Indiana/00; reassortant H1N1 (β cluster), A/swine/Minnesota/02; and human-like (δ cluster), A/swine/Ontario/03. Representative isolates were selected from each cluster and were chosen on the basis of intracluster and intercluster HA sequence variability. One isolate represented the α cluster (α-I). Two isolates each were selected for the β and γ clusters (β-I, β-II, γ-I, and γ-II) and three isolates were selected to represent the δ cluster (δ-I, δ-II, and δ-III).
Figure 1: Amino acid dendrogram of 37 contemporary δ-cluster swine influenza virus isolates used in a study that genetically and antigenically characterized δ-cluster SIVs isolated over a 12-month period. Included are reference strains A/Swine/Ontario/52156/03,12 A/Swine/Illinois/00685/05,13 A/Swine/Minnesota/07002083/07,13 A/Swine/North Carolina/00573/05,13 and A/Swine/Illinois/07003243/07.13 Solid vertical lines indicate subcluster designation. State of origin is indicated in parentheses. Samples originated from North Carolina, Indiana, Ohio, Illinois, Minnesota, Iowa, Oklahoma, and Texas. 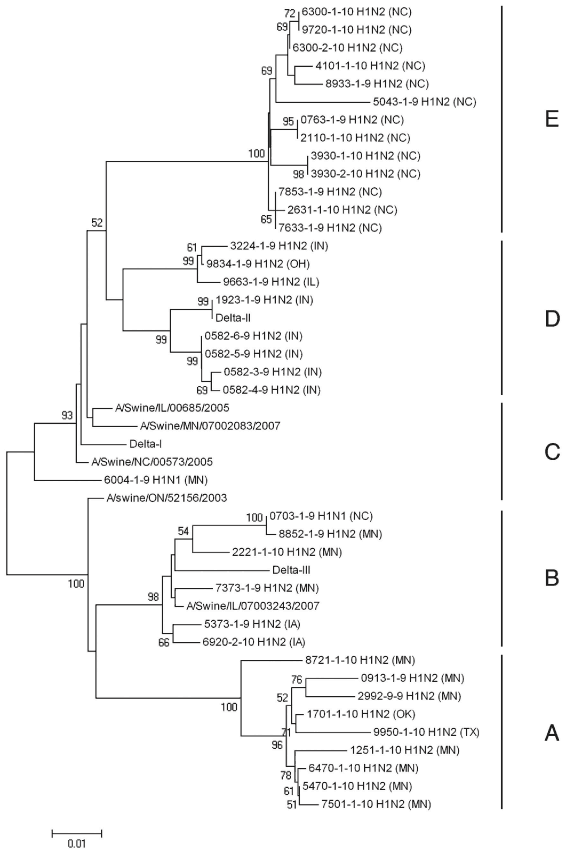 |
Virus isolation was performed in swine testicle (ST) cells grown in Dulbecco’s minimum essential medium (DMEM) containing 5% fetal bovine serum at 37ºC with 5% CO2. For virus propagation, DMEM without fetal bovine serum was used. Viral titers were determined by quantitative reverse-transcription polymerase chain reaction (RT-PCR) and hemagglutination using 0.5% turkey red blood cells in phosphate-buffered saline (PBS).
Swine influenza virus reference isolates were grown in ST cells to a high titer (1280 to 2560 hemagglutination units) and inactivated using 5 mM binary ethyleneimine (BEI) for 24 hours at 37ºC. A 0.1 M stock of BEI was made by combining equal volumes of 0.2 M 2-bromoethylamine hydrobromide (Sigma Aldrich, St Louis, Missouri) and 0.4 N H2SO4 and stirring overnight at room temperature. Vaccines were formulated using killed virus and Trigen, a proprietary adjuvant (Newport Laboratories, Worthington, Minnesota).
Generation of antisera
Thirty-three 3-week-old pigs were obtained from a commercial high-health herd, and 3 days prior to vaccination, whole blood was collected and the SIV-negative status of these pigs was confirmed using a commercial ELISA (FlockChek Avian Influenza MultiS-Screen Antibody Test Kit; Idexx Laboratories Inc, Westbrook, Maine) and by hemagglutination inhibition (HI) and serum neutralization (SN) assays using the eight reference viruses used in this study (Table 1). Nasal swabs were also negative for SIV by real-time RT-PCR.15 Pigs were injected on Days 0, 7, 14, and 21 with 2 mL of vaccine administered intramuscularly. Whole blood was collected on Days 0 and 35. Groups of three pigs were used for antisera generation. Sera were pooled prior to testing. Because sera collected from vaccinated pigs had very high HI titers (homologous HI 1280 to > 20,480), sera were diluted four-fold to 16-fold in serum from cesarean-derived, colostrum-deprived pigs to achieve homologous HI titers of 1280. The diluted reference antisera stocks were aliquoted and frozen at -80ºC. Antiserum was also generated against a commercial SIV vaccine containing a δ-cluster SIV (FluSure XP; Pfizer, New York, New York). To generate antiserum against the commercial vaccine, three pigs were vaccinated on Days 0 and 21 and blood samples were collected on Day 35. Antiserum generated from commercial vaccine was not diluted for serological testing. Homologous HI testing was not performed on commercial-vaccine antiserum, as the SIV isolates used in the product are not publicly available. In addition, negative control antiserum was generated by injecting pigs with ST cell debris and adjuvant. Antiserum generated from the ST cell debris was not diluted.
Table 1: H1 swine influenza virus reference isolates circulating in US swine herds* and used to generate antisera for use in the high throughput serum neutralization assay
* Field strains isolated from clinical samples (nasal swabs or lung tissue) were submitted to Newport Laboratories (Worthington, Minnesota) for isolation and genetic analysis in 2009 and 2010 as part of routine diagnostic testing. † The percent similarity of the isolates to the corresponding commonly used cluster reference strain is shown. Sequence comparisons performed on translated amino acids. |
Serological assays
Cross-reactivity was determined by HI and HTSN assays. All sera were heat inactivated at 56ºC for 30 minutes prior to use. The HTSN assay was performed in duplicate as previously described, with minor modifications.14 Briefly, antisera were diluted 1:800 in DMEM and viruses were diluted in DMEM to 1000 median tissue culture infectious doses (TCID50) per 100 μL as determined by titration on a monolayer of ST cells. The virus and antiserum mixture was incubated at 37ºC for 1 hour, and 200 μL was transferred to a 2-day-old monolayer (100% confluent) of ST cells. Plates were incubated for 4 days at 37ºC with 5% CO2. One hundred microliters of supernatant was transferred to a 96-well U-bottom plate, and 75 μL of 20 μM methyl-umbelliferyl-N-acetyl-neuraminic acid was added to all wells of each plate. Plates were incubated for 2 hours at 37ºC with 5% CO2, and then 100 μL of stop solution (0.1 M glycine, pH 10.7 in 25% ethanol) was added and fluorescence was measured on a fluorescent plate reader with excitation and emission filters of 355 and 460 nm, respectively. Mean fluorescence for the duplicate wells was determined and used to calculate the serum neutralization ratio = (sample – virus-only control) ÷ (no-virus control – virus-only control). Serum neutralization (SN) ratios were calculated for each virus and antiserum combination performed in duplicate, and results were reported as a mean SN ratio. Previous work determined that an SN ratio > 0.31 is positive.14 Specifically, evaluation of 40 SIV isolates against ST antisera using a nonparametric prediction interval with 97.5% confidence established a positive SN ratio threshold of > 0.31. A similar analysis using the minor assay modifications described above supported the cutoff value of > 0.31 (data not shown). High throughput serum neutralization assays were repeated on 2 separate days and values were reported as means.
The HI assay was performed using standard assay procedures as previously described.16 Briefly, sera were first treated with receptor-destroying enzyme for 24 hours at 37ºC. Treated sera were next hemadsorbed with a 20% suspension of washed turkey erythrocytes in PBS for 30 minutes at room temperature. Serial two-fold dilutions of sera were used in the HI assay (1:10 to 1:20,480). Hemagglutination inhibition was assessed using 0.5% washed turkey erythrocytes with viral titers of 4 to 8 hemagglutination units per well. Hemagglutination inhibition titer was determined as the reciprocal of the highest dilution that showed complete inhibition of hemagglutination. Hemagglutination inhibition assays were performed in duplicate and results were reported as arithmetic mean titers.
Genetic analysis
Ribonucleic acid was isolated from ST cell harvest fluids with the 5X MagMAX-96 Viral Isolation Kit (Ambion, Inc, Austin, Texas). Full-length viral RNA segment 4 was amplified by RT-PCR using the OneStep RT-PCR kit (Qiagen, Valencia, California) and previously published primers.12,17 Polymerase chain reaction products and primers were submitted to the University of Minnesota Advanced Genetic Analysis Center for DNA sequencing. Neuraminidase subtyping was performed using published primers in conjunction with the extracted RNA and enzymes described above.18 Deoxyribonucleic acid sequences were translated into amino acids and aligned by pairwise cluster analysis using Bionumerics (Applied Maths, Inc, Austin, Texas). Phylogenetic analyses were performed using MEGA 4.0.19 Evolutionary analyses were performed using the neighbor-joining method, and tree topology was verified with 1000 bootstrap replicates. Swine influenza virus subtype reference strains were included in the analysis: α cluster, A/Swine/Indiana/88; β cluster, A/Swine/Minnesota/02; γ cluster, A/Swine/Indiana/00; δ cluster, A/Swine/Ontario/52156/03, A/Swine/Illinois/00685/05, A/Swine/North Carolina/00573/05, A/Swine/Illinois/07003243/07, and A/Swine/Minnesota/07002083/07.
Results
Genetic analysis
Pairwise cluster analysis of the HA gene of 164 H1 SIV isolates from June 2009 through May 2010 identified 51 δ-cluster isolates. Full-length gene sequences of 37 randomly chosen δ-cluster isolates were determined by nucleotide sequencing, and the sequences were deposited with GenBank under accession numbers HQ896632 to HQ896668. Pair-wise cluster analysis of translated amino acids and phylogenetic analysis of nucleotide sequences identified five subclusters (Figure 1) with 97% to 100% similarity within subclusters and 95% to 97% similarity between subclusters. Isolates used for antisera generation were located in subclusters B, C, and D. Hemagglutinin and NA subtyping found 35 H1N2 and two H1N1 isolates. Isolates in subcluster A originated from Minnesota and Texas. Isolates in subcluster B originated from Minnesota, Iowa, and North Carolina. The sole isolate in subcluster C originated from Minnesota. Indiana, Illinois, and Ohio were the sources of isolates in subcluster D. All subcluster E isolates originated from North Carolina.
High throughput serum neutralization analysis
The 37 δ-cluster isolates were analyzed using a panel consisting of three δ-cluster antisera and the commercial-vaccine antiserum in an HTSN assay (Table 2). The percentage of isolates neutralized by the three H1 antisera varied by subcluster. Neutralization rates were highest for subclusters C (100%, one of one), D (88%, seven of eight) and E (54%, seven of 13). Lower neutralization rates were seen with subclusters A (0%, zero of nine) and B (33%, two of six). The commercial-vaccine antiserum failed to neutralize any viruses used in this study. Overall, only 46% of the isolates were neutralized by at least one of the four antisera generated by δ-cluster isolates used in this study. No neutralization was observed with antiserum generated against the cell line (ST) used to grow SIV.
Table 2: High throughput serum neutralization (HTSN) assay ratios of 37 contemporary H1 δ-cluster swine influenza virus (SIV) isolates*
* The H1 δ-cluster isolates (described in Table 1) were assayed against a panel of antisera generated against isolates representing circulating H1 δ-cluster SIV, swine testicle (ST) cell debris, and a commercial SIV vaccine containing a δ-cluster isolate (FluSure XP; Pfizer, New York, New York). † Isolates are ordered as in the dendrogram in Figure 1. ‡ HTSN ratio calculated as (sample – virus-only control) ÷ (no-virus control – virus-only control). Ratios > 0.31 (positive assays) are shown in red. Average of duplicate analysis is reported.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Reference antisera δ-I and δ-II were cross-reactive with each other in the HTSN assay. Additionally, most isolates neutralized by either δ-I or δ-II antibodies were neutralized by both antisera. All isolates neutralized by δ-I or δ-II antisera were located in subclusters C, D, or E. Reference viruses δ-I and δ-II localized to subclusters C and D, respectively. Other than the homologous virus, reference antiserum δ-III neutralized only two isolates in subcluster B and had weak neutralization activity towards a single subcluster E isolate (9720-1-10) and reference virus δ-II.
Antisera generated against other H1 SIV clusters were used to further characterize the 37 field isolates. Antisera were raised against two γ-cluster, two β-cluster, one α-cluster, and the human pandemic influenza (pH1N1) isolates. None of the δ-cluster isolates were neutralized by α-cluster, β-cluster, or γ-cluster antisera or antiserum generated against pH1N1 (Table 3).
Table 3: High throughput serum neutralization (HTSN) assay ratios of 37 contemporary H1 δ-cluster swine influenza virus (SIV) isolates
* The δ-cluster isolates (described in Table 1) were assayed against a panel of antisera generated against isolates representing circulating H1 SIV β, γ, and α clusters, pandemic H1N1, and swine testicle (ST) cell debris. Isolates are ordered as in the dendrogram in Figure 1. † HTSN assay ratios > 0.31 represent positive assays. Average of duplicate analysis is reported. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Hemagglutination inhibition analysis
Hemagglutination inhibition assay results for a subset of thirteen 2010 isolates and three reference viruses are shown in Table 4. Overall, the HTSN and HI results were in good agreement. Five of the 13 isolates (38%) had positive reciprocal titers (≥ 40) to at least one δ-cluster antiserum. One isolate, 6920-2-10, was negative on the HTSN assay but had HI reciprocal titers of 30 to 160 with the three δ-cluster antisera.
Table 4: High throughput serum neutralization (HTSN) assay ratio and hemagglutination inhibition (HI) antibody titers* of thirteen 2010 δ-cluster swine influenza virus isolates†
* Positive HTSN assay ratios and HI titers are shown in red. Average of duplicate analysis is reported. † Isolates (described in Table 1) were assayed using antisera generated against the three δ-cluster isolates (δ-I, δ-II, and δ-III) and a commercial swine influenza vaccine containing a δ-cluster SIV (FluSure XP; Pfizer, New York, New York). |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Discussion
Following the discovery of SIV with human-like HA genes in Canadian pigs in 2003, triple reassortant SIV with human-like HA soon became endemic in US swine herds. Sequence data (2005 to 2008) from the University of Minnesota Veterinary Diagnostic Laboratory found that approximately 30% of the H1 SIVs contained human-like HA genes.8 Similarly, genetic analysis of SIV isolates over a 12-month period during 2009 to 2010 in the authors’ laboratory identified approximately one-third of H1 isolates as belonging to the δ cluster (unpublished data). Routine antigenic analysis using a panel of H1 SIV antisera frequently failed to neutralize approximately 50% of these isolates, warranting further study. Similarly, others have reported antigenic diversity within the δ cluster.13,20-22
Genetic analysis of 37 recent δ-cluster isolates found five distinct subclusters. Isolates within subclusters were 97% to 100% similar, while subclusters were 95% to 97% similar. Analysis of the 600-base-pair region encompassing the antigenic regions of HA (base pairs 322 to 921) identified five subclusters with 95% to 100% similarity within subclusters and 91% to 95% similarity between subclusters. These results are in good agreement with other genetic analyses reporting approximately 92% amino-acid similarity within the δ cluster.20 In contrast to previous work13 that reported two genetic clusters within δ-cluster viruses, as determined by complete genome sequencing of four isolates, this study identified five distinct subclusters. The larger sample set used in this study (37 viruses) likely explains these differences.
Representative isolates from subclusters B, C, and D, along with contemporary α, β, γ, and pH1N1 viruses, were used to generate hyperimmune antisera. Antigenic characterization of the δ-cluster isolates using the H1 SIV antisera panel in an HTSN assay found no cross-reactivity between δ-cluster isolates and antisera generated against α, β, γ, and pH1N1 viruses, in agreement with previous work.13
The δ-I and δ-II antisera generated from isolates in subclusters C and D were cross-reactive and antigenically similar: δ-I and δ-II antisera neutralized eight of the nine isolates located in these subclusters. Additionally, seven of 13 subcluster E isolates were neutralized by δ-I and δ-II antisera. These results demonstrate that despite amino-acid differences, subclusters C, D, and E are antigenically related and consequently are likely one lineage. These data also suggest that significant antigenic drift has occurred in subclusters D and E, as isolates in these two subclusters were not neutralized by δ-I and δ-II antisera, especially in subcluster E.
Subclusters A and B were genetically and antigenically distinct from one another and from subclusters C, D, and E. Delta-III antiserum, generated against a subcluster B isolate, recognized only two of six subcluster B isolates. Although subcluster B isolates are genetically > 97% similar, substantial antigenic differences exist within the subcluster. Additionally, with one exception, δ-III antiserum failed to neutralize subcluster A, C, D, and E isolates, suggesting that subcluster B is antigenically distinct from the other subclusters and likely results from a separate human-like HA introduction into swine.
Subcluster A isolates were not neutralized by any antisera used in this study. Consequently, subcluster A likely represents another unique lineage of human-like HA introduced into swine herds.
Previous work demonstrated that HTSN assay results were in good agreement with the traditional HI assay.14 A subset of thirteen 2010 isolates were also analyzed by the HI assay to further confirm the HTSN results. Eight isolates were negative and four were positive on both assays. In contrast, isolate 6920-2-10 was negative on the HTSN assay and positive on the HI assay. Hemagglutination inhibition titers for 6920-2-10 ranged from 40 to 160, suggesting that the HI assay has greater sensitivity than the HTSN assay. A serum dilution of 1:800 was previously determined to yield strong homologous (intracluster) and weak intercluster HTSN ratios. This high serum dilution is the likely reason for the lower sensitivity of the HTSN assay.
Antisera generated against a commercial SIV vaccine that contains a δ-cluster isolate failed to neutralize any of the isolates used in this study. As this could be due to lower sensitivity in the HTSN assay, the commercial-vaccine antiserum was analyzed with a subset of isolates in the HI assay. All isolates analyzed had titers < 10 in the HI assay. A likely explanation for the lack of higher titers is that significant antigenic drift has occurred since isolation of the δ-cluster strain used in the commercial product. Alternately, the commercial δ-cluster isolate may represent a different lineage than the three reported in this work consisting of 2009-2010 isolates. Unfortunately, sequence information is not publicly available for the commercial δ-cluster vaccine strain (Dr Marie Gramer, e-mail communication, January 2011).
Geographical analysis of the isolates revealed an association between origin of the isolates and genetic subcluster. The one notable exception was 0703-1-9, which was isolated from a sample from North Carolina. Given the movement of swine in modern farming operations, further surveillance is clearly warranted.
As recently demonstrated by the pandemic H1N1 in humans, influenza virus is constantly changing. Reassortment between viral segments can lead to large-scale antigenic shift and movement between species.11,23 The introduction of wholly human H1N1 and the reassortment between human-like H1N1 and triple reassortant SIV resulted in the formation of δ-cluster isolates. The genetic and antigenic analysis reported in these studies suggests that at least three separate introductions of human-like HA genes have occurred, and descendants of these lineages are currently circulating in US swine herds. Additionally, the antigenic heterogeneity within the five subclusters further suggests that antigenic drift has occurred within the δ cluster. Clinical studies using multiple δ-cluster isolates demonstrated that these viruses induced typical influenza-like disease in experimentally infected animals and were transmissible to contact animals, further illustrating the need to control these viruses.13,21 On the basis of this work, either multiple representatives of this cluster may be required in commercial vaccines, or herd-specific vaccines may be required to protect swine.
Implications
• Significant genetic and antigenic heterogeneity exists in δ-cluster H1 SIV isolates.
• Commercial vaccines containing a single δ-cluster isolate may not protect swine against infection with contemporary influenza viruses.
• Either multiple H1 SIV virus types may be required in commercial vaccines, or herd-specific vaccines may be required to protect swine from infection.
References
1. Vincent AL, Lager KM, Ma W, Lekcharoensuk P, Gramer MR, Loiacono C, Richt JA. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Vet Microbiol. 2006;118:212-222.
2. Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. 2004;103:67–73.
3. Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res. 2002;85:199–210.
4. Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon KJ, Krauss S, Webster RG. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856.
5. Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. Swine influenza viruses: A North American perspective. Adv Virus Res. 2008;72:127–154.
6. Karasin AI, Olsen CW, Anderson GA. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J Clin Microbiol. 2000;38:2453–2456.
7. Karasin AI, Landgraf J, Swenson S, Erickson G, Goyal S, Woodruff M, Scherba G, Anderson G, Olsen CW. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J Clin Microbiol. 2002;40:1073–1079.
*8. Rapp-Gabrielson VJ, Sorensen S, Nitzel G, Wicklund E, Wood MD, Kuhn M, Gramer M. Updating swine influenza vaccines. Proc AASV. San Diego, California. 2008;261–264.
9. Ma W, Gramer M, Rossow K, Yoon KJ. Isolation and genetic characterization of new reassortant H3N1 swine influenza virus from pigs in the Midwestern United States. J Virol. 2006;80:5092–5096.
10. Ma W, Vincent AL, Gramer MR, Brockwell CB, Lager KM, Janke BH, Gauger PC, Patnayak D, Webby RJ, Richt JA. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci USA. 2007;104:20949–20954.
11. Brockwell-Staats C, Webster RG, Webby RJ. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza Other Respi Viruses. 2009;3:207–213.
12. Karasin AI, Carman S, Olsen CW. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003–2005). J Clin Microbiol. 2006;44:1123–1126.
13. Vincent AL, Ma W, Lager KM, Gramer MR, Richer JA, Janke BH. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes. 2009;39:176–185.
14. Hause BM, Oleson TA, Bey RF, Stine DL, Simonson RR. Antigenic categorization of contemporary H3N2 swine influenza virus isolates using a high-throughput serum neutralization assay. J Vet Diagn Invest. 2010;22:352–359.
15. Richt JA, Lager KM, Clouser DF, Spackman E, Suarez DL, Yoon KJ. Real-time reverse transcription-polymerase chain reaction assays for the detection and differentiation of North American swine influenza viruses. J Vet Diagn Invest. 2004;16:367–373.
16. Cottey R, Rowe CA, Bender BS. Influenza virus. Curr Protoc Immunol. May 2001. doi: 10.1002/0471142735.im1911s42.
17. Zou S. A practical approach to genetic screening for influenza virus variants. J Clin Microbiol. 1997;35:2623–2627.
18. Choi YK, Goyal SM, Kang SW, Farnham MW, Joo HS. Detection and subtyping of swine influenza H1N1, H1N2 and H3N2 viruses in clinical samples using two multiplex RT-PCR assays. J Virol Methods. 2002;102:53–59.
19. Tamura K, Dudley J, Nei M, Kumar S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599.
*20. Rapp-Gabrielson VJ, Nitzel GP, Yonkers TK, Wicklund E, Czach JL, Evans JM, Behan SK, Taylor LP, Gramer MR. Antigenic and genetic variations of endemic human-like (delta cluster) swine influenza H1N1 and H1N2 field isolates from the United States. Proc AASV. Omaha, Nebraska. 2010;377–378.
*21. Ciacci-Zanella JR, Vincent AL, Zanella EL, Brockmeier S. Comparison of human-like H1 swine influenza virus isolates. Proc AASV. Omaha, Nebraska. 2010;95–97.
*22. Gramer M, Nitzel G, Wicklund E, Evans J, Kula J, Jeffers J, Taylor L, Ricker T, Rapp-Gabrielson VJ. Serological cross-reactions between reference strains and field isolates representing different genetic clusters of H1N1 and H3N2 swine influenza virus. Proc AASV. San Diego, California. 2008;99–101.
23. Bastien N, Antonishyn NA, Brandt K, Wong CE, Chokani K, Vegh N, Horsman GB, Tyler S, Graham MR, Plummer FA, Levett PN, Li Y. Human infection with a triple reassortant swine influenza A (H1N1) virus containing the hemagglutinin and neuraminidase genes of seasonal influenza virus. J Infect Dis. 2010;201:1178–1182.
* Non-refereed references.


