| Original research | Peer reviewed |
Cite as: Foss DL, Kopta LA, Paquette JA, et al. Identification of Helicobacter suis in pig-producing regions of the United States. J Swine Health Prod. 2013;21(5):242–247..
Also available as a PDF.
SummaryObjectives: To develop a non-culture-based method to determine levels of Helicobacter suis infection in porcine stomachs and to test the method in a sample of pigs from a variety of regions in the United States. Materials and methods: A polymerase chain reaction (PCR) assay was developed to quantitate total Helicobacter generic DNA and Helicobacter suis species-specific DNA in pig stomachs. Primers were derived from 16s ribosomal RNA (rRNA) gene sequences, selected on the basis of relative conservation and divergence of sequences across the various Helicobacter species. The assay was standardized using cloned 16s rRNA sequences and was initially tested with DNA isolated from cultured H suis. Gastric mucosal scrapings were collected from pigs in three geographic regions of the United States, including the North (Minnesota and Michigan), East Central (Iowa), and South (Oklahoma and North Carolina). Results: Of a total of 118 pigs tested, approximately half (55.1%; 95% CI, 46.1%-63.8%) were positive for H suis DNA. Helicobacter suis DNA was detected in pigs from all states tested. Implications: Helicobacter suis is present in US pigs and may be relevant to pig health and production. This quantitative PCR assay will facilitate further study of H suis in pigs, including potential therapeutic and prophylactic interventions. | ResumenObjetivos: Desarrollar un método no basado en cultivo para determinar los niveles de infección de Helicobacter suis en estómagos porcinos y para probar el método en una muestra de cerdos de diversas regiones es Los Estados Unidos. Materiales y métodos: Se desarrolló una prueba de reacción en cadena de polimerasa (PCR por sus siglas en inglés) para cuantificar un DNA genérico de Helicobacter total y el DNA específico de las especies Helicobacter suis en estómagos de los cerdos. Los primers se derivaron de las secuencias del gen RNA ribosomal (rRNA por sus siglas en inglés) 16s, seleccionadas en base a la conservación relativa y a la divergencia de las secuencias a través de varias especies de Helicobacter. La prueba se estandarizó utilizando secuencias de rRNA 16s clonadas y se probó inicialmente con DNA aislado del cultivo de H suis. Se recolectaron raspados de muscosa gástrica de cerdos en tres regiones geográficas de los Estados Unidos, incluyendo el Norte (Minesota y Michigan), el Este Central (Iowa), y el Sur (Oklahoma y Carolina del Norte). Resultados: De un total de 118 cerdos probados, aproximadamente la mitad (55.1%; 95% CI, 46.1%-63.8%) resultaron positivos al DNA del H suis. Se detectó el DNA del Helicobacter suis en cerdos de todos los estados probados. Implicaciones: El Helicobacter suis está presente en cerdos de Los Estados Unidos y puede ser relevante para la producción y salud porcina. Esta prueba cuantitativa de PCR facilitará estudios adicionales del H suis en cerdos, incluyendo intervenciones profilácticas y terapéuticas potenciales. | ResuméObjectifs: Développer une méthode diagnostiques sans culture pour déterminer le degré d’infection par Helicobacter suis dans des estomacs de porc et tester la méthode dans un échantillonnage de porcs provenant de régions variées des États-Unis.Matériels et méthodes: Une épreuve d’amplification en chaîne par la polymérase (PCR) a été développée pour quantifier l’ADN générique total d’Helicobacter et l’ADN spécifique à l’espèce Helicobacter suis dans des estomacs de porc. Des amorces ont été dérivées des séquences de l’ARN ribosomal 16s (ARNr), sélectionnées sur la base de conservation et divergence relative des séquences parmi les différentes espèces d’Helicobacter. L’épreuve a été standardisée en utilisant des séquences clonées d’ARNr 16s et fut testée initialement avec de l’ADN isolé d’H suis cultivé. Des grattages de la muqueuse gastrique ont été prélevés de porcs dans trois régions géographiques des États-Unis, incluant le nord (Minnesota et Michigan), le centre-est (Iowa), et le sud (Oklahoma et Caroline-du-Nord). Résultats: Sur un total de 118 porcs éprouvés, environ la moitié (55,1%; IC 95%, 46,1%-63,8%) étaient positifs pour la présence d’ADN d’H suis. L’ADN d’H suis a été détecté de porcs provenant de tous les états sélectionnés. Implications: Helicobacter suis est présent chez les porcs américains et peut être pertinent en regard de la santé des porcs et de la production porcine. Cette épreuve PCR quantitative facilitera les études futures sur H suis chez les porcs, incluant les interventions thérapeutiques et prophylactiques potentielles. |
Keywords: swine, Helicobacter suis, gastric ulcers
Search the AASV web site
for pages with similar keywords.
Received: January 2, 2013
Accepted: February 25, 2013
Gastric ulcers are a significant problem in grower and finisher pigs. A 2006 survey of US swine farms identified gastric ulcers as a disease problem in almost half of the medium and large production sites.1 Gastric ulcers were also reported as a disease problem in sows, especially in large sites (total sow and gilt inventory > 500). The etiology of gastric ulcers is thought to be a complex interplay of environmental factors, including feed, housing, and concurrent infectious diseases.2 Pathological changes occur in the pars oesophagea, ranging from hyperkeratosis, erosions, and ulceration to frank hemorrhage and death. Gross pathological changes consistent with gastric ulcers are commonly seen at slaughter, where surveys have reported rates ranging from 5% to 100%.2 The economic impact of gastric ulcers stems both from direct impacts on health and growth and from secondary effects, such as increasing particle size of feed to mitigate ulcer risk, which in turn has a detrimental impact on feed efficiency.2
Gastric, Helicobacter-like bacteria have been reported in pigs from various countries for over 20 years. Some examples include Brazil,3,4 Finland,5 France,6 Netherlands,7 Australia,8 and the United States.9,10 The Helicobacter species found in many of these studies is unclear due to methodology variations, inability to isolate and culture the bacteria described, and evolving understanding and nomenclature of Helicobacter species over the time of the studies. It is apparent, however, that Helicobacter-like organisms are commonly present in pigs throughout the world.
Helicobacter suis is a spiral bacterium found in the stomachs of swine. It was first identified by molecular methods in pigs from Belgium11 and has since been successfully cultured in vitro.12 Helicobacter suis, a distinct species of Helicobacter, is identical to Helicobacter heilmannii type 1 found in humans and is not closely related to Helicobacter pylori or the H pylori-like organisms previously reported in experimental swine.13 It is one of the various so-called non-Helicobacter pylori helicobacters (NHPHs) and is the most common NHPH found in humans.14 The possible significance of these NHPHs in human disease has been extensively reviewed.15 Prevalence of H suis appears to be very low prior to weaning, but increases rapidly following weaning and is very high in adult pigs (> 90%)16 and at slaughter (77%).17
The specific causal relationship of H suis with gastric ulcers and related lesions remains unclear. There has been a long-standing correlation of Helicobacter-like bacteria with gastric lesions in pigs. Several studies have shown that pigs with more severe ulcers tended to have higher numbers of helicobacters or a greater likelihood of being colonized by helicobacters.4,10 Since gastric ulcers have a complex etiology, it is not surprising that attempts to experimentally induce gastric ulcers with H suis have generally been inconclusive.18 However, two recent experimental infection studies have shown H suis infection can reduce daily weight gain by 5%19 to 10%.20 Furthermore, H suis-infected pigs were more likely to have ulcerative lesions of the stomach and microscopic signs of gastritis than were noninfected pigs.20
To date, no comprehensive information on the presence of H suis in US pigs has been available. Previous evidence of helicobacters in swine has been noted in the United States,9,10 but these were not isolated or cultured. Because of the association of H suis with the pathogenesis of gastric ulcers and possible zoonotic potential of H suis, we have developed and piloted a molecular assay for H suis in gastric samples. Using this quantitative polymerase chain reaction (PCR) assay, we found considerable amounts of H suis DNA in pigs from several major pig-producing areas of the United States.
Materials and methods
Animal work was conducted in accordance with the Zoetis Institutional Animal Care and Use Committee guidelines, in compliance with local, state, and national regulations, and subject to local ethical review.
Source of pigs and sample collection
Experimental samples were obtained from Oklahoma (24 pigs), Iowa (37 pigs), and North Carolina (nine pigs). Additional samples were tested from commercially sourced pigs from Michigan (38 pigs) and Minnesota (10 pigs) as part of unrelated studies conducted at Zoetis, Kalamazoo, Michigan. All pigs were of market age, except for those from Michigan and Minnesota, which were approximately 10 and 4 weeks of age, respectively. Samples, including punch biopsies and gastric mucosal scrapings, were collected at slaughter or euthanasia and were frozen for transport or storage. One limitation of PCR testing methods is the small sample size (100 mg) used for the assays. In order to get a more representative sample of the stomach, we collected a large area of stomach mucosa (approximately 50 cm2) by scraping, then mixed and sampled from this larger collection area. Initial testing compared punch biopsies with mucosal scrapings from the same stomachs; however, all summary data is from mucosal scrapings.
DNA extraction
Stomach scrapings or biopsy samples (100 mg) were digested with proteinase K at 55°C overnight and stored at -20°C. Total DNA was extracted from the samples using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, California) according to the manufacturer’s directions for extraction of genomic bacterial DNA. The DNA was eluted in 100 µL nuclease-free water and stored at -20°C. For isolation of control DNA from Helicobacter species, bacteria were pelleted by centrifugation and frozen at -20°C. Bacterial DNA was isolated using the Qiagen DNeasy Blood and Tissue kit according to the manufacturer’s directions for isolation of DNA from gram-negative bacteria. Bacterial DNA was stored at -20°C. Initial experiments were conducted using mucosal samples spiked with Helicobacter DNA to confirm the absence of inhibitory substances in the purified DNA.
Quantitative PCR
Quantitative PCR (TaqMan; Life Technologies, Grand Island, New York) was conducted using sequences common to Helicobacter species (Hb) and specific for H suis (Hs) (Table 1). Sequences for the Helicobacter genus-specific primers and probe were derived from the Helicobacter-common region of the 16s ribosomal RNA (rRNA) gene. Helicobacter suis-specific primers were based on H suis 16s rRNA gene sequence regions previously identified as unique to H suis.17 The H suis-specific TaqMan polymerase chain reaction (TM-PCR) probe was designed using the sequences between the primers. The specific primer and probe sequences were selected from a series of sequences based on the initial assay development and optimization. Primers were obtained from Integrated DNA Technology (Coralville, Iowa) and probes from Life Technologies. The TM-PCR was performed using the described primer and probe sets according to manufacturer’s recommendations (25 µL 2× master mix, 0.2 µL of each primer and probe, 5 µL DNA in a 50-µL total volume reaction) using standard TM-PCR cycling conditions (40 cycles, denaturing and annealing temperatures of 95°C and 60°C, respectively). Helicobacter DNA copy number was calculated (in the TaqMan-based program) using a plasmid DNA standard curve generated with cloned 16s rRNA gene sequences (pCR2.1-TOPO vector; Life Technologies) from H suis, using primers H205F and V126R or Hcom1F and Hcom2R (Table 1) for the H suis-specific and Helicobacter genus-specific TM-PCR, respectively. All samples were tested in triplicate, and all standard curves were generated in duplicate. The cycle threshold (Ct) values for the standard curve template DNA (plasmid DNA containing cloned rRNA sequences) were consistent over time, indicating consistency of amplification and stability of the target DNA. A pure culture of H suis, used to determine primer and probe set amplification sensitivity (Figure 1), was created by limiting dilution followed by expansion of the isolated bacterial clone. Purity of the culture was confirmed by culturing for extraneous bacteria. The calculations shown in Figure 1 are based on an assumption of one 16s rRNA gene per bacterial cell as described for the genomic sequence of H suis.21
Table 1: Primer and probe sequences used to measure Helicobacter suis DNA by quantitative polymerase chain reaction (PCR) in the stomachs of pigs from Iowa, North Carolina, Michigan, Minnesota, and Oklahoma*
* Stomach samples (mucosal scrapings or biopsies or both) were collected from a total of 118 pigs. The 38 pigs from Michigan were 10 weeks of age, the 10 pigs from Minnesota were 4 weeks of age, and all others were market age. DNA was isolated and the amount of total Helicobacter generic and Helicobacter suis-specific DNA was determined by quantitative PCR. Quantitation was based on standard curves generated with control plasmid DNA containing selected Helicobacter sequences. † GenBank sequence EF204589.1, source H suis strain HS1, 16s ribosomal RNA. ‡ Primers used to generate sequences cloned into a plasmid for generating standard curves. |
Figure 1: Comparison of primer and probe set amplification with bacterial genome equivalents shows that the Hb TaqMan polymerase chain reaction (TM-PCR) is more sensitive than the Hs TM-PCR. TM-PCRs with either the Helicobacter genus-specific (Hb) or Helicobacter suis-specific (Hs) primers and probes were spiked with total genomic DNA isolated from the indicated number of H suis bacteria (based on a visual count of bacteria from a pure culture of H suis). All samples were tested in triplicate. A standard curve, based on cloned Helicobacter 16s rRNA gene sequences, was used to calculate the genome copy number. R2 values (Pearson correlation; GraphPad Prism 5; GraphPad Software, Inc, La Jolla, California) for the correlations between spiked bacteria DNA and calculated rRNA DNA numbers are indicated for each assay. Axes are graphed in log10. 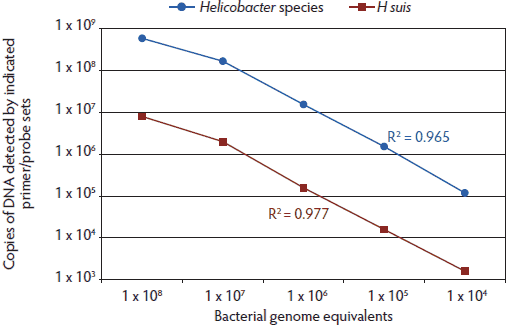 |
Specificities of the primer and probe sets for Helicobacter species and H suis were confirmed by experiments with several species of bacteria, including several Campylobacter jejuni strains, Lawsonia intracellularis, and several Helicobacter species, including H pylori, Helicobacter cynogastricus, Helicobacter felis, and Helicobacter bizzozeronii. The Helicobacter genus-specific TM-PCR detected all of the tested Helicobacter species, while the H suis-specific TM-PCR detected only H suis DNA. Neither primer or probe set detected the non-Helicobacter bacteria (data not shown).
Calculations of percent positive pigs were based on the number of samples positive at any detectable level (> 10 DNA copies per 5-mg sample) versus total samples tested (or total samples tested from a given state). Percentages are shown as 95% confidence intervals (modified Wald method; GraphPad Prism 5).
Results
Standard curves were generated for both the Helicobacter species and H suis primer and probe sets using the respective DNA control plasmids (data not shown). These standard curves were then used to test mock samples that had been spiked with varying amounts of H suis DNA isolated from pure cultures of H suis and corresponding to the indicated number of bacteria (Figure 1). In general, the Hb TM-PCR resulted in higher relative values than the Hs TM-PCR. The differences in relative efficiency of amplification between the standard curve templates (plasmid DNA) and samples (genomic DNA) can be corrected for by using a correlation such as that described in Figure 1, or by using isolated bacterial genomic DNA as the standard curve template. Thus, the Hb TM-PCR is expected to be more sensitive than the Hs TM-PCR; however, the latter would be more specific for H suis and would not detect non-suis Helicobacter species that might be present in test samples.
Initially, both scrapings and punch biopsy samples were tested (samples from Oklahoma and Iowa pigs). We found that the scraping method was more sensitive in detecting Helicobacter DNA. For example, 45.3% (95% CI, 31.0%-61.6%) of the Iowa pig biopsies were positive in the Hb TM-PCR, while 100% (95% CI, 88.8%-100%) of the scraping-derived samples were positive in the same assay. Therefore, all further samples and all data presented here were obtained with the stomach-scraping method.
Helicobacter suis was detected in pigs from all regions of the United States tested. The results are shown as a histogram of all pigs tested having the indicated number of copies of Helicobacter DNA in the stomach mucosal scraping (Figure 2). Helicobacter DNA was detected by the Hb TM-PCR in virtually all samples (from all five states), with some samples having as many as 108 bacteria (DNA equivalents) per 5 mg of gastric mucosal sample (Figure 2A). In the more specific (less sensitive) Hs TM-PCR assay, approximately half of the samples (55.1%; 95% CI, 46.1% 63.8%) were positive and the overall number of bacteria detected was lower (Figure 2B). In this more specific assay, the percent of pigs positive for H suis ranged from 33.4% (95% CI, 21.8%-51.3%) in the Iowa pigs to 100% (95% CI, 67.9%-100%) in the Minnesota pigs, although half of the Minnesota pigs had the lowest detectable numbers of H suis. Among the pigs from Michigan, North Carolina, and Oklahoma, 47.4% (95% CI, 32.5%-62.7%), 56% (95% CI, 26.6%-81.2%), and 79% (95% CI, 64.7%-94.2%) were positive, respectively.
Figure 2: Helicobacter genus-specific or Helicobacter suis-specific DNA in porcine gastric scrapings. TaqMan polymerase chain reactions (TM-PCRs) with either the Helicobacter genus-specific (A) or H suis-specific (B) primers and probes were used to assay total DNA isolated from porcine stomachs. The results are shown as a histogram of the percent pigs from all states that had the indicated number of Helicobacter genome copies per 5 mg of sample. The number of Helicobacter genome copies is graphed in log10. 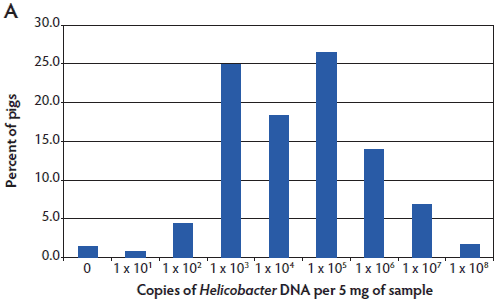 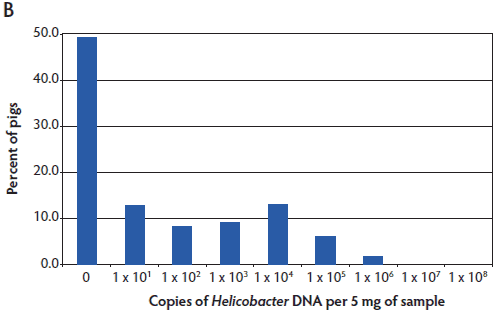 |
Since the currently available H suis genomic sequences are derived from European strains,21 we sequenced several genes from United States-derived H suis (Michigan samples) to obtain preliminary information on the similarity of H suis strains from the United States and Europe. In addition to the similarity of the 16s rRNA sequences, as evidenced by the results of the TM-PCR, we found that both the napA (neutrophil activating protein) and ureA (urease) gene sequences of eight US H suis-containing samples were identical to the published H suis genome (data not shown). Additional methods of analysis (eg, multilocus sequence typing; MLST) could be used to investigate the genetic diversity of H suis within the United States and between US and EU strains without the need for isolation and culture.
Discussion
The role of non-culturable or very fastidious bacteria in disease pathogenesis is likely under-appreciated. The classic example is H pylori, a bacterium observed in human stomachs since the 1880s, but not cultured and confirmed as the cause of gastric ulcers until the 1980s.22-24 Likewise, spiral bacteria have been reported in pig stomachs for many years, but H suis was not isolated and cultured until 2008.12 To date, H suis has been isolated only in Europe, although PCR methods have previously identified Helicobacter sequences consistent with H suis in US pigs.10
In this study, we readily identified and quantitated H suis DNA in pigs from several regions of the United States. Essentially all pigs tested had some detectable Helicobacter DNA. However, fewer samples were positive in the H suis-specific assay. The difference in the results between the two assays could be due to the relative sensitivities of the two primer sets. Spiking experiments with cultured H suis suggests that the H suis-specific assay may underestimate the number of H suis bacteria present in the samples. It is also possible that other Helicobacter species are present in these pigs. Nonetheless, it is clear that some of these pigs had large numbers of H suis in their stomachs (as many as 1 × 106 bacteria per 5 mg of gastric mucosal scraping). The applicability of these results to the overall pig population is very limited due to the small number of samples tested, and is likely confounded by several factors, including age of pigs, time of year samples were collected, and variability in sampling a large surface area. Nonetheless, these results suggest that H suis may be rather common in US pigs.
Previous studies in Europe have found that the incidence of H suis increases with age, starting post weaning and reaching 90% in adult pigs.16 Our results are consistent with H suis being quite common in US pigs; however, this study was not designed to determine estimates of prevalence of H suis in specific states or in pigs of different ages. We found no obvious correlation of pig age with being positive on the TM-PCR for H suis infection, since the young pigs tested from Michigan and Minnesota were very different in the percent of positive pigs – 47.4% (95% CI, 32.5%-62.7%) and 100% (95% CI, 67.9%-100%), respectively. Further studies are needed to address these specific questions.
The apparently high prevalence of H suis raises concerns regarding zoonosis and food safety. Helicobacter suis is the most common NHPH found in humans.14 The potential role of NHPHs from swine (and several other domesticated species) in human disease has been reviewed.15 Evidence for indirect exposure causing H suis infection is lacking, but there has been at least one report of viable H suis in pork samples.25 We have found that H suis spreads rapidly from experimentally infected pigs to adjacent groups of pigs and have identified H suis DNA in saliva from infected pigs (unpublished data). These observations suggest that H suis is highly contagious and easily spread between pigs. Whether H suis is also highly contagious to people in direct contact with pigs is not clear, but close contact with pigs has been associated with H suis infection (known as H heilmannii at the time).26 While other bacteria certainly have more practical significance as pig-derived zoonoses, H suis could well be added to the list. Future studies to confirm transmission of specific, identical H suis strains between pigs and humans are required to address the zoonotic potential of H suis. Further development of assays such as MLST or other DNA fingerprinting methods would enable these studies. One recent study using MLST methods found a close relationship between porcine H suis isolates and a diagnosed human case of H suis infection.27
The relevance of H suis colonization to pig health and production is a matter of ongoing investigation. While it may still be argued that the causal link between H suis colonization and gastric ulcers is circumstantial, it is clear that H suis has the virulence factors to colonize and cause gastric pathology in pigs. The genomes of two H suis strains have been sequenced, and H suis contains an array of virulence and colonization factors required for gastric survival and pathogenesis of gastric lesions.21 Homology of H suis genes to genes known to be related to colonization and virulence of H pylori shows that H suis has numerous genes encoding mechanisms of mucosal adhesion, resistance to low pH, chemotaxis, and motility. Homologs to several H pylori genes related to gastric pathology have also been identified, including vacA (a cytotoxin of gastric epithelial cells), napA, and ggt (γ-glutamyl transpeptidase, an apoptosis-inducing protein).21 Further studies have clearly defined an important and specific role for H suis γ-glutamyl transpeptidase in inducing both necrosis and apoptosis of gastric epithelial cells.28 The pathogenesis of H suis has also been studied in laboratory animal models. Infection of mice or gerbils results in gastritis characterized by parietal cell death, proliferation of epithelial cells, lymphocyte infiltration, and formation of lymphoid follicles.29,30 These lymphoid follicles are sites of an active immune response consisting of B lymphocytes, dendritic cells, and T lymphocytes, mediated in part by secretion of interferon-γ.31 These potential mechanistic links between H suis infection and gastritis leading to ulceration of the pars oesophagea are further supported by recent studies in pigs that linked experimental H suis infection to gastritis.20
Experimental studies of H suis in pigs have begun to ascertain the relevance of H suis colonization to pig health and production. Challenge of pigs with mouse-derived H suis results in colonization and gastritis (infiltration by lymphocytes and plasma cells), but no significant induction of ulcers in the pars oesophagea.18 Experimental infection of pigs has been reported to lower daily weight gain of infected groups by 5%19 to 10%.20 It is likely that gastric ulcers have a complex etiology that makes an experimental challenge model difficult, and it is not yet known if there are growth or feed-efficiency impacts of H suis colonization besides its possible role in causing gastric ulcers. Effective challenge models are expected to be complex and will likely need to include some of the well-known cofactors involved in gastric ulcer formation, such as feed quality and continuity, concurrent disease, and other stressors. Vaccine studies in mice have demonstrated the feasibility of vaccination to prevent H suis colonization32 and disease,33 and antibiotic susceptibility has been evaluated in mice34 and in cultured H suis.35 Thus, vaccine or therapeutic trials are feasible and may define the relevance of H suis to pig health better than extensive efforts to develop a complex challenge model.
Development of assays not dependent on isolation and culture, as we describe here, will enable further studies into the relevance of H suis to pig and human health and the development of strategies for prevention and control of related disease.
Implications
• Detection of H suis DNA in pigs from five states in three geographic areas suggests that H suis is present in the US pig population.
• This quantitative PCR assay will enable further investigation of the relevance of H suis to pig health and production and human health.
Acknowledgments
We thank Dr Richard Ducatelle (Ghent University, Belgium) and colleagues for providing Helicobacter suis strains and technical discussions. Collection of samples was coordinated by Zoetis, Veterinary Operations Pork (Drs Michael J. Kuhn and Alan B. Scheidt). We also thank Dr Scanlon Daniels (Circle H Animal Health, Dalhart, Texas) and Dr Jeff W. Harker (Swine Health Services, Frankfort, Indiana) for providing pig stomachs for testing.
Conflict of interest
Dr Dennis L. Foss, Laurice A. Kopta, Jennifer A. Paquette, Dr Terry L. Bowersock, Dr Leszek J. Choromanski, Dr Jeffery E. Galvin, Traci K. Godbee, and Robert W. Laurinat are employees of Zoetis (formerly Pfizer Animal Health). Margaret Sanchez was an employee of Pfizer Animal Health at the time of the study. Zoetis has a research collaboration and licensing agreement with the University of Ghent, holder of Helicobacter suis-related patents.
References
1. USDA. Swine 2006 Part II: Reference of Swine Health and Health Management Practices in the United States, 2006. Fort Collins, Colorado: USDA. 2007. Available at: http://www.aphis.usda.gov/animal_health/nahms/swine/#swine2006. Accessed 25 April 2013.
2. Friendship R. Gastric ulcers. In: Straw BE, D’Allaire S, Mengeling WL, Taylor, DJ, eds. Diseases of Swine. Ames, Iowa: Iowa State Press; 1999:685–694.
3. Queiroz DM, Rocha GA, Mendes EN, Lage AP, Carvalho AC, Barbosa AJ. A spiral microorganism in the stomach of pigs. Vet Microbiol. 1990;24:199–204.
4. Queiroz DM, Rocha GA, Mendes EN, De Moura SB, De Oliveira AM, Miranda D. Association between Helicobacter and gastric ulcer disease of the pars esophagea in swine. Gastroenterology. 1996;111:19–27.
5. Utriainen M, Hänninen ML. Detection of Helicobacter-like bacteria in porcine gastric biopsy samples by amplification of 16S rRNA, ureB, vacA and cagA genes by PCR. Vet Res Commun. 1998;22:373–383.
6. Cantet F, Magras C, Marais A, Federighi M, Megraud F. Helicobacter species colonizing pig stomach: molecular characterization and determination of prevalence. Appl Environ Microbiol. 1999;65:4672–4676.
7. Roosendaal R, Vos JH, Roumen T, van Vugt R, Cattoli G, Bart A, Klaasen HL, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG. Slaughter pigs are commonly infected by closely related but distinct gastric ulcerative lesion-inducing gastrospirilla. J Clin Microbiol. 2000;38:2661–2664.
8. O’Rourke JL, Solnick JV, Neilan BA, Seidel K, Hayter R, Hansen LM, Lee A. Description of ‘Candidatus Helicobacter heilmannii’ based on DNA sequence analysis of 16S rRNA and urease genes. Int J Syst Evol Microbiol. 2004;54:2203–2211.
9. Melnichouk S, Friendship R, Dewey C, Bildfell R, Smart N. Helicobacter-like organisms in the stomach of pigs with and without gastric ulceration. J Swine Health Prod. 1999;7:201–205.
10. Choi YK, Han JH, Joo HS. Identification of novel Helicobacter species in pig stomachs by PCR and partial sequencing. J Clin Microbiol. 2001;39:3311–3315.
11. De Groote D, van Doorn LJ, Ducatelle R, Verschuuren A, Haesebrouck F, Quint WG, Jalava K, Vandamme P. ‘Candidatus Helicobacter suis’, a gastric helicobacter from pigs, and its phylogenetic relatedness to other gastrospirilla. Int J Syst Bacteriol. 1999;49:1769–1777.
12. Baele M, Decostere A, Vandamme P, Ceelen L, Hellemans A, Mast J, Chiers K, Ducatelle R, Haesebrouck F. Isolation and characterization of Helicobacter suis sp. nov. from pig stomachs. Int J Syst Evol Microbiol. 2008;58:1350–1358.
13. Krakowka S, Ringler SS, Flores J, Kearns RJ, Eaton KA, Ellis JA. Isolation and preliminary characterization of a novel Helicobacter species from swine. Am J Vet Res. 2005;66:938–944.
14. Baele M, Pasmans F, Flahou B, Chiers K, Ducatelle R, Haesebrouck F. Non-Helicobacter pylori helicobacters detected in the stomach of humans comprise several naturally occurring Helicobacter species in animals. FEMS Immunol Med Microbiol. 2009;55:306–313.
15. Haesebrouck F, Pasmans F, Flahou B, Chiers K, Baele M, Meyns T, Decostere A, Ducatelle R. Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin Microbiol Rev. 2009;22:202–223.
16. Hellemans A, Chiers K, De Bock M, Decostere A, Haesebrouck F, Ducatelle R, Maes D. Prevalence of ‘Candidatus Helicobacter suis’ in pigs of different ages. Vet Rec. 2007;161:189–192.
17. De Groote D, Ducatelle R, van Doorn LJ, Tilmant K, Verschuuren A, Haesebrouck F. Detection of “Candidatus Helicobacter suis” in gastric samples of pigs by PCR: comparison with other invasive diagnostic techniques. J Clin Microbiol. 2000;38:1131–1135.
18. Hellemans A, Chiers K, Decostere A, De Bock M, Haesebrouck F, Ducatelle R. Experimental infection of pigs with ‘Candidatus Helicobacter suis’. Vet Res Commun. 2007;31:385–395.
*19. Kumar S, Haesebrouck F, Pasmans F, Flahou B, Dewulf J, Chiers K, Ducatelle R. An experimental Helicobacter suis infection reduces daily weight gain in pigs. Proc IPVS. Vancouver, Canada. 2010;O80:117.
20. De Bruyne E, Flahou B, Chiers K, Meyns T, Kumar S, Vermoote M, Pasmans F, Millet S, Dewulf J, Haesebrouck F, Ducatelle R. An experimental Helicobacter suis infection causes gastritis and reduced daily weight gain in pigs. Vet Microbiol. 2012;160:449–454.
21. Vermoote M, Vandekerckhove T, Flahou B, Pasmans F, Smet A, De Groote D, Van Criekinge W, Ducatelle R, Haesebrouck F. Genome sequence of Helicobacter suis supports its role in gastric pathology. Vet Res. 2011;42:51. doi: 10.1186/1297-9716-42-51.
22. Goodwin CS, Armstrong JA, Marshall BJ. Campylobacter pyloridis, gastritis, and peptic ulceration. J Clin Pathol. 1986;39:353–365.
23. Marshall BJ, Armstrong JA, McGechie DB, Glancy RJ. Attempt to fulfill Koch’s postulates for pyloric Campylobacter. Med J Aust. 1985;142:436–439.
24. Konturek JW. Discovery by Jaworski of Helicobacter pylori and its pathogenetic role in peptic ulcer, gastritis and gastric cancer. J Physiol Pharmacol. 2003;54(suppl 3):23–41.
25. De Cooman LM, Pasmans F, Flahou B, Smet A, Houf K, Ducatelle R, Haesebrouck F. Detection of viable Helicobacter suis in pork by a combination of ethidium monoazide (EMA) and real time-PCR. Helicobacter. 2011;16:142.
26. Meining A, Kroher G, Stolte M. Animal reservoirs in the transmission of Helicobacter heilmannii. Results of a questionnaire-based study. Scand J Gastroenterol. 1998;33:795–798.
27. Liang J, Ducatelle R, Pasmans F, Smet A, Haesebrouck F, Flahou B. Multilocus sequence typing of the porcine and human gastric pathogen Helicobacter suis. J Clin Microbiol. 2013;51:920–926.
28. Flahou B, Haesebrouck F, Chiers K, Van Deun K, De Smet L, Devreese B, Vandenberghe I, Favoreel H, Smet A, Pasmans F, D’Herde K, Ducatelle R. Gastric epithelial cell death caused by Helicobacter suis and Helicobacter pylori gamma-glutamyl transpeptidase is mainly glutathione degradation-dependent. Cell Microbiol. 2011;13:1933–1955.
29. Yamamoto K, Tanaka H, Nishitani Y, Nishiumi S, Miki I, Takenaka M, Nobutani K, Mimura T, Ben Suleiman Y, Mizuno S, Kawai M, Uchiyama I, Yoshida M, Azuma T. Helicobacter suis KB1 derived from pig gastric lymphoid follicles induces the formation of gastric lymphoid follicles in mice through the activation of B cells and CD4 positive cells. Microb Infect. 2011;13:697–708.
30. Flahou B, Haesebrouck F, Pasmans F, D’Herde K, Driessen A, Van Deun K, Smet A, Duchateau L, Chiers K, Ducatelle R. Helicobacter suis causes severe gastric pathology in mouse and Mongolian gerbil models of human gastric disease. PLoS ONE 2010;5(11):e14083. doi: 10.1371/journal.pone.0014083.
31. Mimura T, Yoshida M, Nishiumi S, Tanaka H, Nobutani K, Takenaka M, Suleiman Y, Yamamoto K, Ota H, Takahashi S, Matsui H, Nakamura M, Miki I, Azuma T. IFN-gamma plays an essential role in the pathogenesis of gastric lymphoid follicles formation caused by Helicobacter suis infection. FEMS Immunol Med Microbiol. 2011;63:25–34.
32. Hellemans A, Decostere A, Duchateau L, De Bock M, Haesebrouck F, Ducatelle R. Protective immunization against “Candidatus Helicobacter suis” with heterologous antigens of H. pylori and H. felis. Vaccine. 2006;24:2469–2476.
33. Flahou B, Hellemans A, Meyns T, Duchateau L, Chiers K, Baele M, Pasmans F, Haesebrouck F, Ducatelle R. Protective immunization with homologous and heterologous antigens against Helicobacter suis challenge in a mouse model. Vaccine. 2009;27:1416–1421.
34. Hellemans A, Decostere A, Haesebrouck F, Ducatelle R. Evaluation of antibiotic treatment against “Candidatus Helicobacter suis” in a mouse model. Antimicrob Agents Chemother. 2005;49:4530–4535.
35. Vermoote M, Pasmans F, Flahou B, Van Deun K, Ducatelle R, Haesebrouck F. Antimicrobial susceptibility pattern of Helicobacter suis strains. Vet Microbiol. 2011;153:339–342.
* Non-refereed reference.
