Original research
Peer reviewed
Effect of concurrent infections on persistence and shedding of porcine reproductive and respiratory syndrome virus and transmissible gastroenteritis virus
William L. Mengeling, DVM, PhD; Ronald D. Wesley, DVM, PhD; Kelly M. Lager, DVM, PhD; Ann C. Vorwald; Deborah F. Clouser
WLM, RDW, KML, ACV, DFC: Virus and Prion Diseases of Livestock Research Unit, National Animal Disease Center, USDA, Agricultural Research Service, 2300 Dayton Avenue, PO Box 70, Ames, IA 50010. Tel: 515-663-8254; Fax: 515-663-8458; E-mail: wmengeli@nadc.ars.usda.gov.
Mengeling WL, Wesley RD, Lager KM, et al. Effect of concurrent infections on persistence and shedding of porcine reproductive and respiratory syndrome virus and transmissible gastroenteritis virus. J Swine Health Prod. 2002;10(2):67-73. Also available as a PDF
Summary
Objective: To determine the duration of persistence and shedding of porcine reproductive and respiratory syndrome virus (PRRSV) and transmissible gastroenteritis virus (TGEV) for pigs inoculated with one or both viruses.
Methods: Pigs were allocated to three principal groups (Principal Groups A, B, and C: four pigs per group). They were inoculated with PRRSV (Principal Groups B and C) at 14 +/- 2 days old (Day 0) and with TGEV (Principal Groups A and C) on Day 13. On Day 28 and at or about 14-day intervals (up to and including Day 83), a group of two age-matched, naive pigs were placed in contact with each Principal Group for an interval of 10 days. Samples obtained at selected intervals from both principals and contacts were tested for PRRSV (sera and lung lavage samples) and TGEV (rectal swabs). Sera were also tested for antibody to both viruses.
Results: Contact transmission of PRRSV and TGEV was detected for 56 and 15 days, respectively. A longer interval of infectionwith PRRSV (up to 121 days) was detected when samples from principals were tested using a nested-set polymerase chain reaction. Concurrent infection with PRRSV and TGEV did not enhance clinical effects, shedding, or persistence of eithervirus.
Implications: Pigs infected with PRRSV and TGEV should be kept isolated for more than 8 and 2 weeks, respectively, to reduce the chance for virus transmission. Concurrent infection with TGEV and PRRSV is likely to have little or no effect on subsequent shedding or persistence of either virus.
Keywords: swine, transmissible
gastroenteritis virus, porcine reproductive and respiratorysyndrome
virus, concurrent infection,persistent infection
swine, transmissible
gastroenteritis virus, porcine reproductive and respiratorysyndrome
virus, concurrent infection,persistent infection
Received: May 15, 2001
Accepted: August 16, 2001
Porcine reproductive and respiratory syndrome (PRRS)1 and transmissible gastroenteritis (TGE)2 are two of the most economically important diseases affecting swine production in the United States. Both are caused by enveloped RNA viruses that, in young pigs, ofteninduce severe and sometimes fatal illness. Because of the high prevalence of PRRS virus (PRRSV) and TGE virus (TGEV) in the swine population in the United States, 3-5 concurrent infections are likely, especially in endemically infected herds.
The most common means by which PRRSV and TGEV are disseminated within and among swine herds is believed to be shedding from acutely or persistently infected pigs. Consequently, the most effective way to prevent the continuous circulation of either virus is to isolate infected pigs until shedding subsides, presumably as the result of a protective immune response. Although this approach is straightforward in principle, it is also often costly. Therefore, it is important to determine the likely duration of persistence and shedding and to limit isolation to no longer than necessary.
Numerous studies have already addressed the issues of persistence and transmission of PRRSV6-11 and TGEV.12-15 However, our knowledge of these issues is still incomplete, especially in regard to PRRSV. Moreover, the potential confounding effect of concurrent infection has yet to be defined.
In the study reported here, we experimentally addressed the temporal aspects of persistenceand contact transmission of PRRSV and TGEV by first infecting pigs (principals) with either or both viruses. Then, at approximately 14-day intervals, naive pigs (contacts) were placed in direct contact with principals. To determine the intervals of persistence and transmission, principals and contacts were tested for virusand homologous antibody.
Materials and methods
Experimental design
The experiment included 42 pigs: 12 principals and 30 contacts. Principals comprised three groups (A, B, and C) of four pigs per group that were members of four litters (ie, one pig of each litter per group). Pigs of Principal Groups B and C were each inoculated intranasally with 2 mL of cell culture medium containing 6 x 106 median cell culture infective units of PRRSV (virulent strain NADC-8)16 when they were 14 +/- 2 days of age (designated Day 0 of the experiment). About 2 weeks later (Day 13), pigs of Principal Groups A and C were each inoculated intragastrically via stomach tube with 5 mL of cell culture medium containing 1.8 x 105 plaque-forming units of TGEV (virulent strain Miller).17 Serum samples were obtained from all Principal Groups on Days 0 (just before inoculation with PRRSV) 7, 13 (just before inoculation with TGEV), 20, 28, 38, 53, 66, 80, 94, 108, and 121. Rectal swab samples were obtained from all Principal Groups on Days 13 (just before inoculation with TGEV), 14, 15, 16, 17, 18, 20, and 23 (ie, during an interval of 10 days after inoculation with TGEV). Lung lavage fluids (containing large numbers of alveolar macrophages and hereafter referred to as lavage samples) were obtained from all Principal Groups on Days 38, 53, 66, 80, 94, and 121.
On Day 28 (ie, 28 days after Principal Groups B and C had been inoculated with PRRSV and 15 days after Principal Groups A and C had been inoculated with TGEV), blood samples were obtained from six age-matched pigs (Contact Group I) which were then placed in contact with each of the three Principal Groups (two Contact Group I pigs per Principal Group). After 10 days of contact, each of the Principal Groups was moved to another isolation room (each group to a separate room). On the same day, blood and lavage samples were obtained from all principals and contacts. Sampling was performed in a separate isolation area to preclude the possibility of contaminating isolation rooms with potentially virus-containing fluids. Four days later (Day 42), six additional age-matched pigs (Contact Group II) were put in contact with the principals (two Contact Group II pigs per Principal Group). After 6 more days (Day 48, and 20 days after initial contact with Principal Groups), blood was obtained from all Contact Group I pigs. Contact Group I pigs were then euthanized and lavage samples were obtained. This sequence of procedures was repeated a total of five times, with contact of two Contact Group pigs with each Principal Group beginning on Days 28, 42, 56, 70, and 83 (ie, approximately 4, 6, 8, 10, and 12 weeks after inoculation of Principal Groups B and C with PRRSV and approximately 2, 4, 6, 8, and 10 weeks after inoculation of Principal Groups A and C with TGEV). All pigs were observed at least twice daily for clinical signs.
Pigs
All principals were from litters farrowed in isolation rooms at the National Animal Disease Center (NADC) by gilts purchased from a specific pathogen free herd. The 12 principals were randomly selected except for litter distribution as previously indicated. All contacts were from litters farrowed off-site at about the same time as the principals and by gilts of the same herd. All contacts were delivered to the NADC on the day they were put in contact with principals (ie, six contacts were delivered to the NADC on each of Days 28, 42, 56, 70, and 83). All gilts and pigs had titers <4 for PRRSV by indirect immunofluorescence18 (IFA) and titers <4 for TGEV by virus serum neutralization24 (SN) on the day they were first used in the experiment.
Sample testing
All serum and lavage samples obtained from principals and contacts were tested for PRRSV by virus isolation directly in MARC-145 cells18 (serum samples) and by cocultivation of alveolar macrophages and MARC-145 cells19,20 (lavage samples). All serum and lavage samples obtained from principals on Day 28 or later were also tested directly or indirectly for PRRSV by genome amplification (open reading frame (ORF) 5 and short contiguous sequences) using a reverse transcriptase, nested-set polymerase chain reaction (PCR).21 The PCR (developed several years after the remainder of the study was completed) was performed with samples that had been kept at -70°C. All were aliquots of the same samples tested for infectious virus except for lavage samples obtained on Days 38 and 121. These had not been saved and PCR was performed with coculture fluids (which had been saved) corresponding to each of these samples, ie, an aliquot of the medium from the previous test for infectious virus. A total of 24 PCR-positive samples (including all PCR-positive samples obtained on Day 53 or later) were also tested by restriction fragment length polymorphism (RFLP) analysis.21,22 The repeatability (reliability) of positive PCR results with samples that were presumed to contain very small amounts of PRRSV (ie, samples that were obtained more than 13 weeks after initial infection and were virus isolation negative) was determined by testing multiple aliquots of the same samples. Namely, both PCR and corresponding RFLP analyses were repeated four or more times with all PCR-positive samples (ie, samples positive the first time tested) that had been obtained on Days 94, 108, and 121. The number of repeats depended primarily on the amount of available sample. All rectal swabs were tested for TGEV by virus isolation.23 All serum samples obtained from principals and contacts were also tested for antibody to PRRSV by IFA18 (titers >=4 were recorded as positive) and for antibody to TGEV by SN24 (titers >=4 were recorded as positive).
Evidence for virus transmission
Evidence for the transmission of PRRSV from principals to contacts was its isolation from serum or lavage samples, or both, from contact pigs. Evidence for the transmission of TGEV from principals to contacts was seroconversion of contact pigs.
Results
Principals
Clinical signs (PRRS). Principal Groups B and C consumed less feed and were less vigorous compared to Principal Group A for several days during the 2 weeks post inoculation of Principal Groups B and C with PRRSV. However, none of the PRRSV-inoculated pigs showed marked clinical signs, and all appeared completely normal by Day 13 when Principal Groups A and C were inoculated with TGEV. Principal group A remained clinically normal prior to its inoculation with TGEV
Clinical signs (TGE). Principal groups A and C became anorectic and developed a watery, gray, fetid diarrhea beginning 2 days post inoculation with TGEV and lastingfor 5 to 7 days. Principal Group B had normal stools and remained clinically normal during the same time interval. All pigs of Principal Groups A and C recovered and appeared clinically normal by 14 days post inoculation with TGEV.
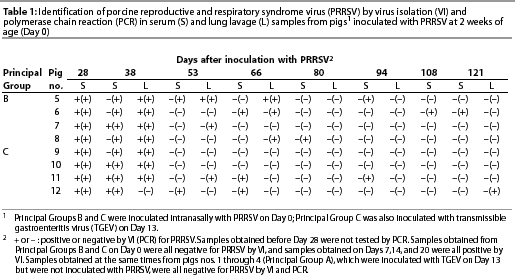
Isolation of PRRSV. All pigs of Principal Groups B and C were viremic on Days 7, 13, 20, and 28. On Day 38, some serum samples (four of eight) and most lavage samples (seven of eight) obtained from the same pigs yielded PRRSV. Thereafter, PRRSV was isolated from only one pig, namely, lavage samples obtained from a pig of Principal Group B on Days 53 and 66 (Table 1). None of the serum or lavage samples obtained at the same times from Principal Group A yielded PRRSV, and none of the serum samples obtained from any of the pigs (Principal Groups A, B, and C) just before inoculation of Principal Groups B and C with PRRSV yielded PRRSV.
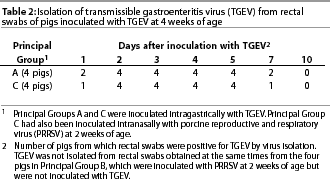
Isolation of TGEV. In all of the eight pigs of Principal Groups A and C, TGEV was isolated from feces (rectal swabs) at some time post inoculation with TGEV. It was detected in the feces of three of the eight pigs on post-inoculation (PI) day 1 (ie, 1 day after inoculation with TGEV), in the feces of all eight pigs on PI days 2 through 5, and in the feces of three of the eight pigs on PI day 7. It was not detected in feces obtained from any of the same pigs just before inoculation with TGEV or on PI day 10 (Table 2). It was never detected in the feces obtained at the same times from pigs of Principal Group B.
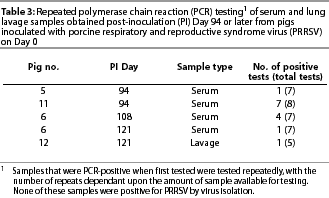
Polymerase chain reaction (PRRSV). On Day 28, ie, the earliest time at which samples were tested for PRRSV by PCR, all serum samples of Principal Groups B and C were positive. On Day 38, all serum samples and seven of eight lavage samples of Principal Groups B and C were PCR-positive. Thereafter, a lesser number of samples obtainedat each time interval (from pigs of Principal Groups B and C) were positive (Table 1). Most (five of six) of the PCR-positive samples obtained on Day 80 or later were serum. In all of the 24 PCR-positive samples that were tested by RFLP analysis, the RFLP pattern was the same as that of the strain with which the pigs had been inoculated. Inconsistent results were obtained with initially PCR-positive samples that had been obtained on Day 94 or later and tested four or more additional times (Table 3). None of the serum or lavage samples obtained from Principal Group A (inoculated with TGEV alone) were PCR-positive.
Antibody detection (PRRSV). All pigs of Principal Groups B and C seroconverted to PRRSV by Day 13 and remained seropositive for PRRSV throughout the remainder of the experiment. All pigs of Principal Group A remained seronegative for PRRSV throughout the experiment.
Antibody detection (TGEV). All pigs of Principal Groups A and C seroconverted to TGEV by Day 20 (7 days after inoculation with TGEV) and remained seropositive for TGEV throughout the remainder of the experiment. All pigs of Principal Group B remained seronegative for TGEV throughout the experiment.
Contacts
Isolation of PRRSV. All but one of the 12 pigs placed in contact with pigs of Principal Groups B and C on Days 28 (Contact Group I), 42 (Contact Group II), and 56 (Contact Group III), became infected with PRRSV. Conversely, none of the eight pigs of Contact Groups IV and V placed in contact with Principal Groups B and C on Days 70 and 83 became infected with PRRSV. None of the ten pigs placed in contact with Principal Group A became infected with PRRSV.
Antibody detection (PRRSV). With one exception, antibody for PRRSV was detected in all of the 11 contact pigs shown to be infected with PRRSV by virus isolation. The single exception was a pig of Contact Group II that was not shown to be infected by virus isolation until Day 20 after initial contact.
Antibody detection (TGEV). Antibody for TGEV was detected only in the two pigs of Contact Group I that contacted Principal Group A on Day 28 (15 days after Principal Group A had been inoculated with TGEV).
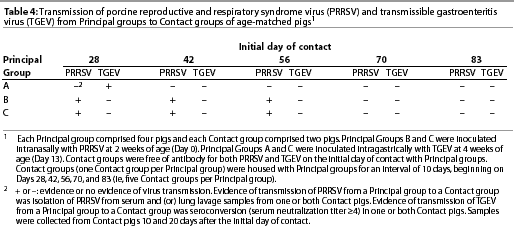
Contact transmission (PRRSV and TGEV). The temporal aspects of contact transmission identified by virus isolation (PRRSV) and antibody detection (PRRSV and TGEV) are summarized in Table 4.
Discussion
Both TGEV and PRRSV were transmitted from infected pigs (principals) to naive pigs (contacts) placed in the same isolation room. However, the duration of virus persistence and shedding was markedly different for the two viruses. Whereas TGEV was isolated from principals for as long as 7 days and transmitted from principals to contacts for as long as 15 days, PRRSV was isolated from principals for as long as 66 days, transmitted from principals to contacts for as long as 56 days, and detected by PCR in serum and lavage samples obtained from principals for as long as 121 days. There was no evidence that concurrent infection,which spanned an interval of at least 7 days and paralleled a severe clinical episode of TGEV-induced diarrhea (Principal Group C), enhanced the persistence, shedding, or clinical manifestations of eithervirus. The actual interval of concurrent infection may have been considerably longer in that it seems unlikely that pigs of Principal Groups A and C would have varied more than a few days in the duration of infection with TGEV. Notice (Table 4) that on the basis of contact transmission, pigs of Principal Group A were still shedding TGEV on Day 28, ie, 15 days after inoculation with TGEV.
The relatively long interval of PRRSV shedding emphasizes the importance of strict and long-term isolation of infected pigs in any attempt to interfere with the persistence of PRRSV in herds in which naive pigs are introduced periodically. Moreover, one might speculate that 56 days is a minimum interval for PRRSV transmission in pigs of the age used in this study, because relatively few pigs (dictated by the need for strict isolation and intensive sampling) were included in either the Principal or Contact Groups. We assume that in much larger groups, such as those typifying commercial swine production, some pigs might shed PRRSV for an even longer period of time, and the number of random contacts with the potential for virustransmission would increase accordingly. The possibility of transmission of PRRSV after even longer intervals is also emphasized by reports of others, namely, the identification of pigs disseminating PRRSV at 9925 and 11210 days after initial infection. In addition, viral RNA, and presumably infectious PRRSV, was identified by PCR in the semen of a boar that had been initially inoculated with PRRSV 92 days earlier.26 The same presumption that any PCR-positive sample is likely to contain infectious virus suggests that even if PRRSV were no longer being shed in secretions or excretions it might be iatrogenically or otherwise transmitted through contactwith contaminated blood. The prevalence and epidemiological impact of these events under farm conditions remains to be determined.
Amplification of the virus genome by PCR was clearly the most sensitive method to identify PRRSV persistence. However, the sporadic nature of test results with samples obtained after the acute stage of infection raises a question in regard to the reliability of PCR for detecting persistently infected pigs. No pig was consistently PCR-positive during the interval beginning on Day 53 and ending on Day 121 (Table 1). Moreover, repeated testing of initially PCR-positivesamples obtained on Day 93 and later (all of which were negative by virus isolation) yielded inconsistent results (Table 3). We assume that these samples contained very little virus, and as a result, some aliquots that were tested contained insufficient virus or viral genome to result in a positive PCR test. For the same reason, it is likely that there would have been additional PCR-positive results had all PCR-negative samples (Table 1) from Principal Groups B and C been tested repeatedly.
From the practical perspective of eliminating PRRSV from an individual herd, there is some solace in the likelihood that contact transmission is of shorter duration than virus persistence. However, even if true, this is likely to be no more than a relative concept in that other studies have suggested that the duration of persistence, and presumably shedding and contact transmission, depends in part on the age of the pig at the time of initial infection.9,10,27
The use of RFLP analysis of PCR-positive samples provided a simple, rapid, and relatively inexpensive method to help exclude the possibility of laboratory contamination. Clearly, this additional step provides support for experimental results such as those reported here. Just as importantly, it has potential diagnostic application. Notably, failure to detect a false-positive test, eg, as a result of cross contamination with the positive control (which is typically run with each PCR procedure), or with another sample being tested concurrently, could have costly repercussions. Although RFLP analysis is most developed for ORF 522 (the ORF amplified in this study) rather than ORF 728 (the ORF most often used for diagnostic PCR), there is apparently no difference in their relative sensitivity in regardto virus detection.21
This study provided relatively few serum and lavage samples for comparison. However, it appeared that at progressively longer times after infection (Table 1; Days 38, 53, and 66), lavage samples provided the better source of infectious PRRSV, an observation consistent with our previous studies,16,18,20 and with the commonly held belief that alveolar macrophages are the primary site of PRRSV replication in pigs.1 However, PCR testing revealed a potentially different story in that after Day 66, most (five of six) of the PCR-positive samples were serum (Table 1). Assuming that these data are representative, there is a possibility that the predominant site or sites of PRRSV replication change with time. This could explain why others have been so successful in detecting persistent infection of tonsils,10,29-31 whereas our previous, shorter-term (<=10 weeks) comparisons of tonsil specimens, serum samples, and lavage samples for PRRSV identification by virus isolation indicated a clear advantage for the lavage samples.16,18
The ages and times at which pigs were inoculated with PRRSV and TGEV in this study were selected on the basis of what we believed would allow the most meaningful, sensitive, and reliable evaluation of our experimentalobjectives. First, the use of weaned pigs (ie, at or about 2 weeks of age) allowed us to exclude potential litter variation by including a member of each of the four litters in each experimental group. At the same time, it allowed us to use pigs that more closely reflect farm conditions than if we had used either gnotobiotic or caesarian-derived, colostrum-deprived pigs. Second, we knew that inoculation of pigs with PRRSV when they were 2 weeks old was likely to result in a prolonged, nonlethal infection that would extend beyond the clinical manifestations of subsequent TGEV infection. Third, we knew that inoculationof pigs with the virulent Miller strain of TGEV when they were at or about 4 weeks of age was likely to result in a severe diarrhea from which pigs would recover - and yet it was near the time (<=2 weeks of age) when pigs would usually succumb to infection. Therefore, we hypothesized that any enhancement of clinical illness, or virus persistence, or virus shedding, or some combination of these, caused by either TGEV or PRRSV, would be more likely and more obvious under these conditions than if the same pigs had been inoculated with TGEV when they were either younger or older. Fourth, we knew from previous observations that by 2 weeks after infection of young pigs with PRRSV (about the time pigs of this study were inoculatedwith TGEV), there would likely be a marked, PRRSV-induced lymphoid response (as evidence by generalized enlargement of lymph nodes) which might, in turn, reflect a progressive immune dysfunction. This idea was supported by the fact that littermates inoculated with the same strain of PRRSV (as part of a separate study) at the same time and in the same manner, and then necropsied 15 days later, had markedly enlarged lymph nodes as well as extensive lung lesions.32 Therefore, it was our belief that if there were any measurable effects of combined infection with PRRSV and TGEV, they would be most pronounced if young pigs were inoculated first with PRRSV.
For TGEV, the only difference in clinical signs, transmission, and viral persistence between Principal Groups A (infected with TGEV alone) and C (infected with both TGEV and PRRSV) was that contact transmission of TGEV was detected on Day 28 (15 days after inoculation with TGEV) for Principal Group A, but not for Principal Group C (Table 4). This might have been due to an enhanced immune response as a result of previous inoculation with PRRSV,33 but more likely it is simply a result of random variation. For PRRSV, there was no clear difference between Principal Groups B (infected with PRRSV alone) and C (infected with both TGEV and PRRSV) in respect to any of the parameters measured. Therefore, we conclude that neither virus measurably altered the effects of the other. We emphasize, however, that this study was completed, except for PCR testing, before the emergence of highly virulent strains of PRRSV associated with atypical PRRSV.34,35 It may, therefore, reflect only what might be expected with strains of PRRSV of virulence similar to that of strain NADC-8, used to inoculate the Principals.
We assume that the eventual cessation of viremia and detectable shedding of PRRSV was due primarily to a slow but progressive development of immunity capable of preventing, or at least minimizing, continued virus replication. However, in addition to the temporal aspects of the immune response,it is likely that age played a role. Older pigs are likely to be viremic for a shorter interval and presumably shed less virus.25 This difference seems to be especially pronounced when the duration of infection of adults is compared with that of congenitally infected pigs.9,27 Older pigs, as well as adults, may also have a higher threshold for the establishment of infection. If so, the greater age of both principals and contacts at each subsequent contact time would have influenced our results in regard to the perceived interval of shedding. Nevertheless, we believe that the experimentaldesign based on age-matched contacts is the most meaningful in regard to typical farm conditions wherein mixing of pigs at any stage of the production cycle most often involves contact of pigs at or near the same age.
Implications
- Pigs initially infected within about the first 4 weeks of age can shed PRRSV and TGEV in amounts resulting in contact transmission for at least 56 days and 15 days, respectively. These intervals should be exceeded when isolation times are being selected for strategies to eliminate TGEV and PRRSV from infected herds.
- The interval of persistent infection with PRRSV may be appreciably longer than the interval of contact transmission.
- The nested-set PCR is a highly sensitive test to identify persistent infection with PRRSV. However, a positive test does not ensure contact transmission and a negative test does not ensure absence of infection.
- Restriction fragment length polymorphism analysis is a simple, ancillary tool that can be used to help exclude laboratory contamination as a cause of a positive PCR and thus increase test reliability.
- An additional and severe clinical insult, such as that caused by TGEV during dual infection of young pigs with TGEV and PRRSV, is likely to have little or no effect on subsequent shedding or persistence of PRRSV, and concurrent infection with PRRSV is likely to have little or no effect on subsequent shedding and persistence of TGEV.
Disclaimer
No endorsements are herein implied. Brand names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standards of the products, and the use of the names by the USDA implies no approval of the products to the exclusion of others that may also be suitable.
References - refereed
1. Benfield DA, Collins JE, Dee SA, Halbur PG, Joo HS, Lager KM, Mengeling WL, Murtaugh MP, Rossow KD, Stevenson GW, Zimmerman JJ. Porcine reproductive and respiratory syndrome. In: Straw BE, D'Allaire S, Mengeling WL, Taylor DJ, eds. Diseases of Swine. 8th ed. Ames, Iowa: Iowa State University Press. 1999:201-232.
2. Saif LJ, Wesley RD. Transmissible gastroenteritis and porcine respiratory coronavirus. In: Straw BE, D'Allaire S, Mengeling WL, Taylor DJ, eds. Diseases of Swine. 8th ed. Ames, Iowa: Iowa State University Press. 1999:295-325.
5. Wesley RD, Woods RD, McKean JD, Senn MK, Elazhary Y. Prevalence of coronavirus antibodies in Iowa swine. Can J Vet Res. 1997;61:305-308.
8. Albina E, Madec F, Cariolet R, Torrison J. Immune response and persistence of the porcine reproductive and respiratory syndrome virus in infected pigs and farm units. Vet Rec. 1994;134:567-573.
11. Allende R, Laegreid WW, Kutish GF, Galeota JA, Wills RW, Osorio FA. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J Virol. 2000;74:10834-10837.
12. Morin M, Morehouse LG, Solorzano RF, Olson LD. Transmissible gastroenteritis in feeder swine: role of feeder swine in the epizootiologic features. Am J Vet Res. 1974;35:251-255.
13. Underdahl NR, Mebus CA, Torres-Medina A. Recovery of transmissible gastroenteritis virus from chronically infected experimental pigs. Am J Vet Res. 1975;36:1473-1476.
14. Woods RD, Wesley RD. Transmissible gastroenteritis coronavirus carrier sow. Adv Exp Med Biol. 1998;440:641-647.
15. Pensaert MB, Haelterman EO, Burnstein T. Transmissible gastroenteritis of swine: virus-intestinal cell interactions. I. Immunofluorescence, histopathology and virus production in the small intestine through the course of infection. Arch Gesamte Virusforsch. 1970;31:321-334.
16. Mengeling WL, Vorwald AC, Lager KM, Brockmeier SL. Comparison among strains of porcine reproductive and respiratory syndrome for their ability to cause reproductive failure. Am J Vet Res. 1996;57:834-839.
17. Wesley RD, Woods RD, Correa I, Enjuanes L. Lack of protection in vivo with neutralizing monoclonal antibodies to transmissible gastroenteritis virus. Vet Microbiol. 1988;18:197-208.
18. Mengeling WL, Lager KM, Vorwald AC. Diagnosis of porcine reproductive and respiratory syndrome. J Vet Diagn Invest. 1995;7:3-16.
20. Mengeling WL, Vorwald AC, Lager KM, Brockmeier SL. Diagnosis of porcine reproductive and respiratory syndrome using infected alveolar macrophages obtained from live pigs. Vet Microbiol. 1996;49:105-115.
21. Umthun AR, Mengeling WL. Restriction fragment length polymorphism analysis of strains of porcine reproductive and respiratory syndrome virus by use of a nested-set reverse transcriptase-polymerase chain reaction. Am J Vet Res. 1999;60:802-806.
23. Wesley RD, Woods RD. Induction of protective immunity against transmissible gastroenteritis virus after exposure of neonatal pigs to porcine respiratory coronavirus. Am J Vet Res. 1996;57:157-162.
24. Woods RD, Wesley RD, Kapke PA. Neutralization of porcine transmissible gastroenteritis virus by complement-dependent monoclonal antibodies. Am J Vet Res. 1988;49:300-304.
26. Christopher-Hennings J, Nelson EA, Nelson JK, Swenson SL, Zimmerman JJ, Chase CL, Yager MJ, Benfield DA. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of adult boars. J Vet Diagn Invest. 1995;4:456-464.
27. Mengeling WL, Lager KM, Vorwald AC. Temporal characterization of transplacental infection of porcine fetuses with porcine reproductive and respiratory syndrome virus. Am J Vet Res. 1994;55:1391-1398.
28. Christopher-Hennings J, Nelson EA, Nelson JA, Hines RJ, Swenson, SL, Hill HT, Zimmerman JJ, Katz JB, Yaeger MJ, Chase CCL, Benfield DA. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J Clin Microbiol. 1995;33:1730-1734.
32. Wesley RD, Mengeling WL, Lager KM. Prior infection of nursery-age pigs with porcine reproductive and respiratory syndrome virus does not affect the outcome of transmissible gastroenteritis virus challenge. J Vet Diagn Invest. 1998;10:221-228.
34. Zimmerman J, Epperson W, Wills RW, McKean JD. Results of the recent survey of the membership of the AASP for outbreaks of sow abortion and mortality syndrome. Swine Health Prod. 1997;5:74-75.
35. Mengeling WL, Lager KM, Vorwald AC. Clinical consequences of exposing pregnant gilts to strains of porcine reproductive and respiratory syndrome (PRRS) virus isolated from field cases of "atypical" PRRS. Am J Vet Res. 1998;59:1540-1544.
References - non refereed
3. USDA:APHIS:VS:CEAH. Prevalence of PRRS virus in the United States. National Animal Health Monitoring System (NAHMS) Swine '95 Fact Sheet. Technical Report. USDA:APHIS:VS:CEAH:NAHMS 1997:N225.197.
4. USDA:APHIS:VS:CEAH. Morbidity/mortality & health management of swine in the U.S. National Animal Health Monitoring System (NAHMS) Swine Survey 1989-1990. Technical Report. USDA: APHIS:VS:CEAH:NAHMS 1991:N101.11917.
7. Terpstra C, Wensvoort G, Van Leengoed LAMG. Persistence of Lelystad virus in herds affected by porcine epidemic abortion and respiratory syndrome. Proc IPVS. The Hague, The Netherlands. 1992:118.
9. Benfield DA, Christopher-Hennings J, Nelson EA, Rowland RR, Nelson JK, Chase CL, Rossow KD, Collins JE. Persistent fetal infection of porcine reproductive and respiratory syndrome (PRRS) virus. Proc AASP. Quebec City, Quebec, Canada. 1997:455-458.
10. Benfield DA, Nelson JK, Rossow KR, Nelson C, Steffen M, Rowland RR. Diagnosis of persistent or prolonged porcine reproductive and respiratory syndrome virus infections. Proc 3rd Symp PRRS and Aujeszky's Dis. Ploufragan, France. 1999:151-152.
19. Mengeling WL, Lager KM, Vorwald AC. Diagnosis of porcine epidemic abortion and respiratory syndrome (PEARS). Proc IPVS. Bangkok, Thailand. 1993:56.
22. Wesley RD, Mengeling WL, Andreyev V, Lager KM. Differentiation of vaccine (strain RespPRRS(r)) and field strains of porcine reproductive and respiratory syndrome virus by restriction enzyme analysis. Proc AASP. Nashville, Tennessee. 1996:141-143.
25. Zimmerman J, Swenson SL, Wills RW, Pirtle EC, Yoon K-J, Hill HT, McGinley MJ. Transmission of PRRS virus. Proc Allen D Leman Swine Conf. St. Paul, Minnesota. 1993:51-52.
29. Wills RW, Zimmerman JJ, Yoon K, Swenson SL, McGinley MJ, Hill HT, Platt KB. Porcine reproductive and respiratory syndrome virus: isolation from chronically infected swine. Proc AASP. Omaha, Nebraska. 1995:387-389.
30. Wills R, Zimmerman J, Yoon K-J, Swenson S, McGinley M, Hill H, Platt K, Christopher-Hennings J, Nelson E. Porcine reproductive and respiratory syndrome virus - a persistent infection. Proc 2nd Int Symp Porcine Reproductive and Respiratory Syndrome (PRRS). Copenhagen, Denmark. 1995:19.
31. Horter DC, Pogranichnyy R, Chang C-C, Yoon K-J, Zimmerman JJ. Evaluation of current PRRS diagnostics in detecting persistently infected animals. Proc AASV. Nashville, Tennessee. 2001:503.
33. Molitor TW, Leitner G, Choi CS, Risdahl J, Rossow KD, Collins JE. Modulation of host immune responses by SIRS virus. AASP Newslet. 1992;4:27
