| Brief communication | Peer reviewed |
SummarySalmonella presence, populations, serotypes, and antibiotic susceptibilities in untreated and treated swine manure were determined for farms implementing environmentally superior waste-treatment technologies. The waste-treatment systems surveyed showed potential in reducing Salmonella populations. | ResumenSe determinaron la presencia, población, serotipos, y susceptibilidad a antibióticos de la Salmonella en excretas de cerdo tratadas y no tratadas provenientes de granjas que implementan tecnologías medioambientalmente superiores de tratamiento de desechos. Los sistemas de tratamiento de desechos investigados mostraron potencial para reducir las poblaciones de Salmonella. | ResuméLa présence, les populations, les sérotypes, et les patrons de sensibilité aux antibiotiques d’isolats de Salmonella provenant d’échantillons de lisier de porc non-traités et traités ont été déterminés pour des fermes mettant en place des technologies de traitement du lisier supérieures d’un point de vue environnemental. Les systèmes de traitement étudiés ont démontré du potentiel pour réduire les populations de Salmonella. |
Keywords: swine, Salmonella, waste management, manure
Search the AASV web site
for pages with similar keywords.
Received: June 11, 2010
Accepted: August 27, 2010
Swine manure is generally used as fertilizer and applied to fields for growing agricultural commodities. Salmonella and other pathogens have frequently been isolated from swine wastes.1-3 Land application of Salmonella-contaminated manure may pose an environmental risk if movement occurs to surface and groundwaters.4-6 Sustainable swine production requires the development of innovative and cost-effective waste-management systems to address these and other environmental concerns.7
Much emphasis has been placed on odor and air-quality issues when developing new swine waste-management technologies.8,9 However, the presence, populations, and diversity of Salmonella in the liquid- and solid-waste effluents from different waste-treatment technology systems has received less attention. Thus, the objectives of this field survey were to evaluate the effects of several promising swine waste-management and treatment technologies on reducing Salmonella presence and populations and to characterize the diversity of Salmonella isolates recovered from the waste streams using serotyping and antibiotic-susceptibility analysis.
Materials and methods
In this field study, five environmentally superior waste-treatment technologies were evaluated for Salmonella presence, populations, serotypes, and antibiotic susceptibilities. Technologies included ambient temperature anaerobic digester, solid separation constructed wetland, up-flow biofiltration, multi-step biological and chemical, and high-solids anaerobic digester (HSAD) treatment systems,6,7 as well as a traditional lagoon treatment system on a conventional swine-production farm. Each treatment technology was implemented on a single swine-production site and was operational for a limited period of time; thus, each farm was sampled only once between August 2002 and March 2003. At the time of sampling, each technology had been in operation between 3 and 6 months.
The ambient temperature anaerobic digester treatment system was designed to “close the loop” with regard to the on-farm processing of swine wastes (Figure 1). This farm was a farrow-to-wean swine operation with approximately 4000 sows in six houses: two farrowing houses and four gestation houses. A pit-recharge system was used to first collect the manure from the farrowing and gestation swine houses. The ambient digester system was installed with an impermeable cover over an in-ground digester that was used for primary waste treatment, with the effluent discharged into one end of a storage pond. The methane biogas produced in the digester was used for generating electricity. Moreover, the ammonia generated in the anaerobic effluent was oxidized to nitrate in the first biofilter. The nitrified water was stored in a tank and then recycled to recharge the pits inside the swine rearing houses. Tomato plants housed in adjacent greenhouses utilized a significant portion of the nutrients recovered from the storage pond after further oxidation of ammonia to nitrate in the second biofilter. In this system, a composite sample was collected from each of the following: fresh feces recovered from the farrowing and gestation pigs, effluent streams exiting the farrowing and gestation swine houses, the ambient digester, the first biofilter, the west storage pond, the second biofilter, the liquid-storage tank in the tomato greenhouse, and the medium used for growing the tomato plants (Figure 1).
Figure 1: Ambient temperature anaerobic digester waste-treatment system installed on a six-house farrow-to-wean swine operation housing 4000 sows. Each sample-collection point (•) represents a composite sample that was analyzed for Salmonella presence and most probable number population estimates. 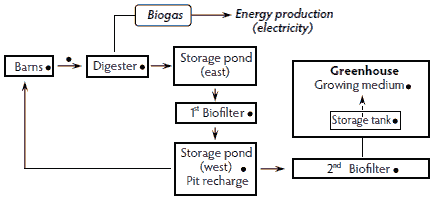 |
The solid separation constructed wetland treatment system consisted of house effluent that was initially pumped to a mechanical solids separator (Figure 2). The liquid was then pumped into a settling basin before flowing into either of two parallel constructed wetlands (inner cell and outer cell) and then to a storage pond. Periodically, settled solids were pumped from the settling basin back to the separator. Some of the treated liquid from the storage pond was used to recharge the house pits. Excess liquids from the storage pond and excess solids from the solids separator were land applied to cropland. The constructed wetland system was designed to convert ammonia contained in the house effluent to nitrate, which was subsequently utilized by the wetland’s cattails. The system was a simple, low-energy alternative to a conventional anaerobic lagoon system. It was installed on a 3520-head swine finishing facility composed of four swine houses. A composite sample was collected from each of the following: fresh swine feces, house effluent, separated solids, inner and outer cell wetland influent, inner and outer cell wetland effluent, and the storage pond (Figure 2).
Figure 2: Solid separation constructed wetland waste-treatment system installed on a four-house finishing swine operation housing 3520 finisher pigs. Each sample-collection point (•) represents a composite sample that was analyzed for Salmonella presence and most probable number population estimates. 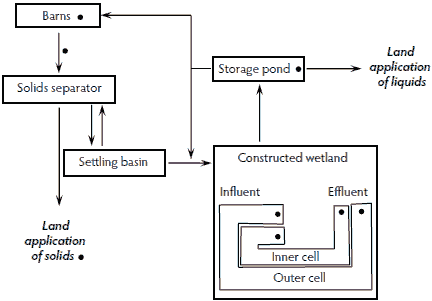 |
The up-flow biofiltration treatment system incorporated solids separation and aerobic biological treatment of the flushed pre-screened liquid manure for the purpose of reducing chemical oxygen demand, odor, and ammonia emission by promoting nitrification (Figure 3). Five finishing barns with a total of 4300 pigs were connected to the system. Separated solids were pumped into the anaerobic lagoon. Following solids separation, the liquid waste stream was pumped into an equalization tank to allow settling, and then the liquid was pumped upwards into the sequential and aerated biofilters. Plastic fixed media contained within the series of up-flow biofilters provided sufficient surface area for the formation of a bacterial biofilm that aerobically stabilized the organics and converted ammonia to nitrate. The treated effluent was then pumped into the anaerobic lagoon for barn-pit recharge or recirculated into the equalization tank via the solids separator. The biofilters were periodically cleaned through air agitation and a backwash procedure. It should be noted that this waste treatment technology treated only a portion of the total waste generated on the farm. There were other barns on the farm where the wastes were treated conventionally with an anaerobic lagoon system. On this farm, the anaerobic lagoon that received manure from the barns was partitioned by plastic curtains into three sections (L1, L2, and L3), with one section (L3) much larger than the other two; the relative surface areas were 13%, 16%, and 71% of total area, respectively. The L3 section received manure from barns not connected to the treatment system. The L2 section received overflow from the solids separation basin, separated solids, and backwashed biosolids removed from the biofilters. The L1 section received the treated effluent from the biofilters. A composite sample was collected from each of the following: fresh swine feces, house effluent, separated solids, separated liquids, the equalization tank, biofilter effluent, biofilter backwash, the solids reservoir (L2), liquid storage (L1), and the lagoon (L3) (Figure 3).
Figure 3: Up-flow biofiltration waste-treatment system installed on a five-house finishing swine operation housing 4300 finisher pigs. Each sample-collection point (•) represents a composite sample that was analyzed for Salmonella presence and most probable number population estimates. 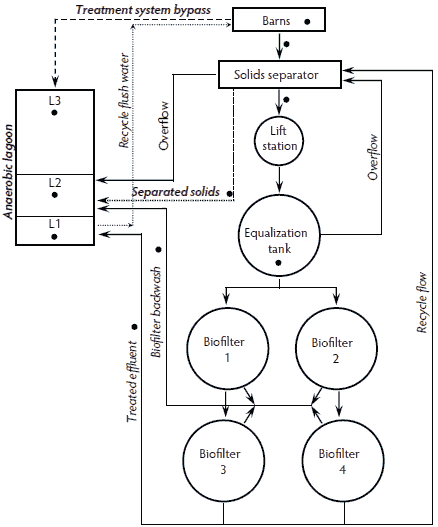 |
The multi-step biological and chemical treatment system incorporated solids separation, with the liquid waste stream subjected to nitrification, de-nitrification, and phosphorus extraction processes (Figure 4). The system was installed on a 4400-head finishing facility composed of six swine houses. First, flushed manure was pumped into a homogenization tank and then separated in the solid-liquid separation module. The liquid-waste stream flowed through nitrification and de-nitrification tanks and then to a settling tank where it was further treated for phosphorus precipitation to calcium phosphate. Treated effluent was stored in the existing lagoon prior to land application. The complete treatment consisted of solids, nitrogen, and phosphorus removal. The separated solids were then subjected to an HSAD treatment system, where swine waste organics were converted to methane biogas. The solids were further composted for conversion to a value-added fertilizer product. For the multi-step biological and chemical treatment system, a composite sample was collected from each of the following: fresh swine feces, house effluent, the homogenization tank, the solid-liquid separation module, the nitrification and de-nitrification tanks, the settling tank, treated effluent, and calcium phosphate fertilizer. For the HSAD treatment system, a composite sample was collected from each of three effluent ports of the HSAD (Figure 4).
Figure 4: Multi-step biological and chemical waste-treatment system installed on a six-house finishing swine operation housing 4400 finisher pigs. Separated solids were subjected to a high-solids anaerobic digester waste-treatment system. Each sample-collection point (•) represents a composite sample that was analyzed for Salmonella presence and most probable number population estimates. 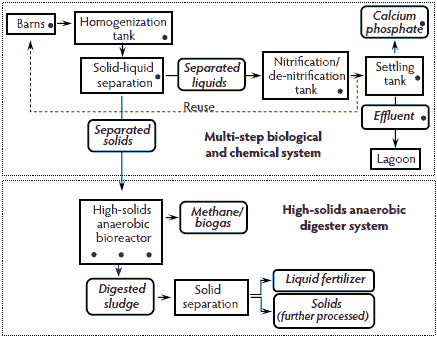 |
The conventional swine-production system consisted of one finishing farm employing a traditional lagoon treatment system. The four-house farm housed 5000 pigs, and the wastes were discharged by flushing to an anaerobic lagoon. A composite sample was collected from each of the following: fresh swine feces, house effluent, and the lagoon.
Solid and liquid samples were aseptically collected in sterile plastic bottles. Following collection, all samples were stored on ice in a transport cooler and processed in the laboratory within 2 hours. Twenty-five-gram solid samples and 25-mL liquid samples were used to determine Salmonella presence and most probable number (MPN) population estimates using culture methods in Rappaport-Vassiliadis broth (Oxoid Ltd, Ogdensburg, New York) and modified lysine iron agar (Oxoid Ltd) as described by Santos et al.10 The three-tube MPN technique employed serial dilution in triplicate tubes. Both culture methods were plated in duplicate. Suspect colonies were confirmed using triple sugar iron agar (Difco, Lawrence, Kansas) and poly-O antiserum (Difco) as previously described.10
Fifty Salmonella isolates were successfully serotyped, originating from the ambient temperature anaerobic digester treatment system (four), the constructed wetland treatment system (nine), the up-flow biofiltration treatment system (18), the multi-step biological and chemical treatment system (12), the HSAD treatment system (two), and the conventional swine operation unit (five). One isolated colony was randomly picked from each sample plate, transferred onto tryptic soy agar (Difco) slants, grown overnight at 37ºC, and submitted to the United States Department of Agriculture National Veterinary Service Laboratories in Ames, Iowa, for serotype determination. Serotyping was based on the Kauffmann-White scheme.
The antimicrobial susceptibilities of the 50 serotyped Salmonella isolates were determined using the Sensititre Susceptibility System (TREK Diagnostic Systems Inc, Westlake, Ohio) as previously described by Santos et al.11 Antimicrobials selected for these assays reflect the recommendations of the National Committee for Clinical Laboratory Standards.12 The susceptibility tests contained the following 15 antimicrobial agents (dilution ranges are indicated in parenthesis): amikacin (0.5 to 64 μg per mL), amoxicillin-clavulanic acid (1 to 32 and 0.5 to16 μg per mL, respectively), ampicillin (1 to 32 μg per mL), cefoxitin (0.5 to 32 μg per mL), ceftiofur (0.12 to 8 μg per mL), ceftriaxone (0.25 to 64 μg per mL), chloramphenicol (2 to 32 μg per mL), ciprofloxacin (0.015 to 4 μg per mL), gentamicin (0.25 to 16 μg per mL), kanamycin (8 to 64 μg per mL), nalidixic acid (0.5 to 32 μg per mL), streptomycin (32 to 64 μg per mL), sulfisoxazole (16 to 256 μg per mL), tetracycline (4 to 32 μg per mL), and trimethoprim-sulfamethoxazole (0.12 to 4 and 2.38 to 76 μg per mL, respectively).
Results
Presence, populations, and serotypes of Salmonella determined from the waste-treatment technologies and the conventional swine-production system are summarized in Table 1. Salmonella populations in the final treated liquid waste streams of the ambient temperature anaerobic digester, constructed wetland, and multi-step biological and chemical treatment systems were under the detection limit (1 log MPN per mL), while in the final treated liquid wastes of the HSAD and the up-flow biofiltration treatment systems, populations were above the detection limit. Salmonella populations in the separated solids wastes from the constructed wetland, multi-step biological and chemical, and up-flow biofiltration treatment systems were also above the detection limit (1 log MPN per g). For each treatment technology, Salmonella populations from fresh swine feces were either low or below the detection limit.
Table 1: Presence, populations, and serotypes of Salmonella recovered from five environmentally superior waste-treatment technologies and a conventional swine-production farm employing a traditional lagoon system*
* Each treatment technology was implemented on a single commercial swine-production site and was operational for a limited period of time. Each farm was sampled once and each sampling point represents a composite sample. Composite samples were used to determine Salmonella presence and most probable number (MPN) population estimates using culture methods in Rappaport-Vassiliadis broth and modified lysine iron agar. The three-tube MPN estimation technique employed serial dilution in triplicate tubes. Both culture methods were plated in duplicate. Presence or absence results are reported as positive (Pos) or negative (Neg) for Salmonella. Log MPN results are reported as the base10 logarithm of the MPN of Salmonella detected per g or mL of sample. The minimum detection limit for the MPN procedure was 10 cells/g or mL of sample (1 log MPN/g or mL). Serotyping was based on the Kauffmann-White Scheme. All Salmonella serotype Typhimurium isolates were classified as var Copenhagen. NA = not applicable. Salmonella was not recovered by either procedure. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
A total of nine serotypes were identified, including Salmonella Derby (15 of 50 isolates, 30%), Salmonella Typhimurium (var Copenhagen, 12 of 50, 24%), Salmonella Johannesburg (8 of 50, 16%), Salmonella Anatum (5 of 50, 10%), Salmonella Infantis (3 of 50, 6%), Salmonella Muenchen (3 of 50, 6%), Salmonella Senftenberg (2 of 50, 4%), Salmonella Heidelberg (1 of 50, 2%) and Salmonella Worthington (1 of 50, 2%).
The antimicrobial agents to which Salmonella isolates (n = 50) were most commonly resistant were tetracycline (29 of 50 isolates, 58%), streptomycin (28 of 50, 56%), ampicillin (10 of 50, 20%), chloramphenicol (6 of 50, 12%), trimethoprim-sulfamethoxazole (3 of 50, 6%), and kanamycin (3 of 50, 6%). Sixty-eight percent of the isolates were resistant to ≥ 1 antimicrobial agent. No Salmonella isolates were resistant to amikacin, amoxicillin-clavulanic acid, cefoxitin, ceftriaxone, ceftiofur, ciprofloxacin, gentamicin, nalidixic acid, or sulfisoxazole.
Discussion
Salmonella populations throughout the waste-treatment systems were similar to those that Vanotti et al3 reported when testing a pilot multi-step biological and chemical treatment system. The initial spike in Salmonella populations following the fresh fecal sampling point may have been due to flushing the feces out of the house with water, thus creating a more favorable environment for Salmonella proliferation. Most treatment technologies showed potential in reducing Salmonella populations in treated liquid manure.
According to the Centers for Disease Control National Salmonella Surveillance System Annual Summary for 2006,13 S Typhimurium was the most frequently reported serotype from human clinical sources. Salmonella Infantis, Muenchen, and Heidelberg were also among the 14 most frequently reported serotypes in 2006. Additionally, the United States Department of Agriculture Food Safety Inspection Service recently reported that S Derby, S Typhimurium (var Copenhagen), S Johannesburg, S Infantis, and S Anatum were among the five most commonly isolated serotypes from swine-processing establishments for calendar year 2009.14 These published statistics are in agreement with the profile of Salmonella serotypes isolated in the present study. Similar to the findings of this study, Salmonella serotypes with multiple-drug resistance characteristics have been previously isolated from production animals and foods of animal origin.15,16
This multiple-farm survey provides an initial assessment of populations, serotypes, and antibiotic resistance of Salmonella recovered from environmentally superior swine waste-treatment technologies which would benefit future risk-assessment studies directed at these waste-management practices. Limitations of this study include single sampling of each site due to the brevity of the evaluation period, and variations in the production stage (finisher pigs versus sows) evaluated across the different waste-treatment technologies. Future pathogen research should include repeated sampling of swine waste-treatment technologies across seasons.
Implications
• The technologies surveyed appear promising for reducing swine-manure Salmonella populations.
• Further study over an extended time frame is warranted prior to drawing any conclusions about the efficacy of these waste-treatment technologies in reducing Salmonella populations.
References
1. Hutchison ML, Walters LD, Avery SM, Synge BA, Moore A. Levels of zoonotic agents in British livestock manures. Let Appl Microbiol. 2004;39:207–214.
2. Hutchison ML, Walters LD, Avery SM, Munro F, Moore A. Analyses of livestock production, waste storage, and pathogen levels and prevalences in farm manures. Appl Environ Microbiol. 2005;71:1231–1236.
3. Vanotti MB, Millner PD, Hunt PG, Ellison AW. Removal of pathogen and indicator microorganisms from liquid swine manure in multi-step biological and chemical treatment. Bioresource Tech. 2005;96:209–214.
4. Baloda SB, Christensen L, Trajcevska S. Persistence of a Salmonella enterica serovar Typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl Env Microbiol. 2001;67:2859–2862.
5. Guan TY, Holley RA. Pathogen survival in swine manure environments and transmission of human enteric illness – A review. J Environ Qual. 2003;32:383–392.
6. Williams M. Development of environmentally superior technologies in the US and policy. Bioresource Tech. 2009;100:5512–5518.
7. Williams M. Development of Environmentally Superior Technologies per Agreements Between the Attorney General of North Carolina and Smithfield Foods, Premium Standard Farms and Frontline Farmers. Phase I Technology Determination Report. 2004. Available at: http://www.cals.ncsu.edu/waste_mgt/smithfield_projects/smithfieldsite.htm. Accessed 23 November 2010.
8. Powers WJ. Odor control for livestock systems. J Anim Sci. 1999;77:169–176.
9. Zahn JA, Hatfield JL, Laird DA, Hart TT, Do YS, DiSpirito AA. Functional classification of swine manure management systems based on effluent and gas emission characteristics. J Environ Qual. 2001;30:635–647.
10. Santos FBO, Li X, Payne JB, Sheldon BW. Estimation of most probable number Salmonella populations on commercial North Carolina turkey farms. J Appl Poult Res. 2005;14:700–708.
11. Santos FBO, D’Souza DH, Jaykus L, Ferket PR, Sheldon BW. Genotypes, serotypes, and antibiotic resistance profiles of Salmonella isolated from commercial North Carolina turkey farms. J Food Prot. 2007;70:1328–1333.
12. National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; Twelfth informational supplement M100-S12. Wayne, Pennsylvania: NCCLS; 2002.
13. CDC. Salmonella Surveillance: Annual Summary, 2006. US Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Georgia. 2008. Available at: http://www.cdc.gov/NCIDOD/DBMD/phlisdata/salmtab/2006/SalmonellaAnnualSummary2006.pdf. Accessed 23 November 2010.
14. USDA, FSIS. Serotypes Profile of Salmonella Isolates from Meat and Poultry Products January 1998 through December 2009. US Department of Agriculture, Food Safety and Inspection Service, Washington, DC. 2009. Available at: http://www.fsis.usda.gov/PDF/
Serotypes_Profile_Salmonella_2009.pdf. Accessed 23 November 2010.
15. Rabatsky-Ehr T, Whichard J, Rossiter S, Holland B, Stamey K, Headrick ML, Barrett TJ, Angulo FJ, The NARMS Working Group. Multidrug-resistant strains of Salmonella enterica Typhimurium, United States, 1997–1998. Emerg Infect Dis. 2004;10:795–801.
16. Gebreyes WA, Altier C. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J Clin Microbiol. 2002;40:2813–2822.
