| Brief communication | Peer reviewed |
Cite as: Harmon KM, Abate SA, Chriswell AJ, et al. Comparison of two commercial real-time reverse transcriptase polymerase chain reaction assays for porcine reproductive and respiratory syndrome virus. J Swine Health Prod. 2012;20(4):184–188.
Also available as a PDF.
SummarySpecimens were tested for porcine reproductive and respiratory syndrome virus by real-time reverse transcriptase polymerase chain reaction (RT-PCR) with two commercial agent-specific assays. Result discrepancy between the two tests was confirmed by conventional RT-PCR, sequencing, or both on six of 423 clinical cases. Neither assay detected every positive case. | ResumenSe examinaron muestras contra el virus del síndrome reproductivo y respiratorio porcino mediante la prueba de reacción en cadena de la polimerasa de transcriptasa reversa (PCR-RT) en tiempo real con dos pruebas comercias agente-específicas. Las discrepancias entre las dos pruebas se confirmaron mediante el PCR-RT convencional, secuenciación, ó ambos en seis de los 423 casos. Ninguna de las dos pruebas detectó todos los casos positivos. | ResuméDes échantillons ont été testés pour le virus du syndrome reproducteur et respiratoire porcin par réaction d’amplification en chaîne par la polymérase utilisant la transcriptase réverse (RT-PCR) avec deux épreuves spécifiques d’agent qui sont disponibles commercialement. La non-concordance des résultats entre les deux épreuves a été confirmée par RT-PCR conventionnelle, par séquençage, ou par les deux techniques, pour six des 423 cas cliniques. Aucune des deux épreuves ne détecta tous les cas positifs. |
Keywords: swine, PCR, PRRSV
Search the AASV web site
for pages with similar keywords.
Received: September 28, 2011
Accepted: February 27, 2012
Porcine reproductive and respiratory syndrome virus (PRRSV) is an RNA virus causing reproductive failure in sows and respiratory disease in piglets and growing pigs.1 The high economic impact of this agent dictates the need for rapid and accurate diagnosis. Polymerase chain reaction (PCR) testing has been increasingly used as a diagnostic test because of its sensitivity and specificity and also because of the ability to complete testing the same day the sample is received. For any PCR test to be effective for determining the absence or presence of the agent of interest, it is essential that it target a highly conserved region of the genome. This is somewhat problematic with RNA viruses such as PRRSV which have high mutation rates, resulting in rapid evolution and genetic variability.2 Even minor mismatches between the PCR target region and the primers and probes used in real-time PCR can cause false-negative results.3-5 Most real-time assays include one primer pair and a single probe for each target, but to enhance the ability to detect a genetically diverse population, it has become more common to utilize multiple primers and probes in a single multiplexed assay. In this study, we compared performance of two commercially available PRRSV-specific real-time reverse transcriptase PCR (RT-PCR) assays for detection and differentiation of North American (NA) and European (EU) subtypes of the virus. The assays are manufactured by Applied Biosystems, Foster City, California (AB), and Tetracore, Rockville, Maryland (TC). The VetMax NA and EU PRRSV reagents (AB) and VetAlert NA and EU PRRSV PCR reagents (TC) each contain multiple primers and probes to detect both NA and EU subtypes. The previous version of the AB reagents (TaqMan NA and EU PRRSV reagents) missed or weakly detected some contemporary PRRSV strains (observed and confirmed in Iowa State University Veterinary Diagnostic Laboratory, 2010). The primers and probes were recently updated to improve the assay’s detection capability. In the current study, the TC and updated AB assays were performed on a subset of clinical specimens submitted to the Iowa State University Veterinary Diagnostic Laboratory to compare the performance of the two commercially available tests.
Materials and methods
The samples used in this study were randomly selected from routine case submissions to our laboratory and consisted of serum (n = 380), lung tissue (n = 277), oral fluid (n = 245), environmental samples (n = 8), nasal swabs (n = 7), semen (n = 6), bronchoalveolar lavage fluid (n = 5), fetal thoracic fluid (n = 4), and assorted tissues (n = 2). Except where noted, nucleic acid extraction was performed using a MagMax Viral RNA Isolation Kit (Life Technologies, Carlsbad, California) in conjunction with a Kingfisher or Kingfisher 96 instrument (Thermo Scientific, Waltham, Massachusetts), according to the manufacturer’s recommendations. For tissue samples, a 10% to 20% (w/v) homogenate was prepared using Earle’s balanced salt solution and then processed on high speed for 120 seconds (Stomacher 80; Seward, Bohemia, New York). The homogenized sample was centrifuged at approximately 4200g for 10 minutes and the supernatant was used in the extraction procedure. Extraction of oral fluids, semen, and environmental samples was performed as described by Chittick et al6 (extraction A1) except the elution buffer volume was reduced to 50 µL. The program used for Kingfisher extraction was the Standard MagMax program provided by Applied Biosystems. Real-time RT-PCR was performed on nucleic acid extracts of a subset of samples submitted to the Iowa State University Veterinary Diagnostic Laboratory using both VetAlert NA and EU PRRSV PCR reagents (TC) and VetMax NA and EU PRRSV PCR reagents (AB). Reactions were performed according to manufacturers’ instructions using the AB 7500 Fast instrument. For each group of samples, both assays were performed on the same day from the same nucleic-acid extract. For the AB testing, analysis was performed using the auto baseline setting. Thresholds for NA and EU PRRSV were set at 0.10 and 0.05, respectively. For TC, manual baseline was used for analysis at the instrument’s default setting of three to 15 cycles. Thresholds for NA and EU PRRSV were set at 51,000 and 145,000, respectively. Forty cycles was used as the cutoff for a positive result for both assays. To monitor for and safeguard against contamination, multiple negative extraction controls, using water as sample, as well as no-template controls, using water as template in the PCR reaction, were included in every 96-well PCR reaction plate. If any of the negative controls produced a cycle threshold (Ct; defined as the cycle at which the amplification curve crosses the threshold) before the end of the run, the samples from the entire plate were retested. A discrepant case was defined as one that exhibited a positive result (Ct ≤ 40) on one or more samples with one assay, and all negative results with the other assay. Nineteen discrepant cases (of a total of 27 cases) were chosen for follow-up and were retested with both assays. Cases that were chosen for follow-up were generally those with earlier Cts (stronger positives) and sufficient volume remaining for additional testing. In cases where pooled sera were originally tested, individual serum samples from the discrepant pools were tested if sufficient sample was available. Cases that repeated as discrepant by real-time RT-PCR were subjected to conventional RT-PCR using primers specified by Christopher-Hennings et al7 and the Qiagen OneStep RT-PCR Kit (Qiagen Inc, Valencia, California), following the manufacturer’s recommendations. Primers were added at a concentration of 600 nM each, and the following thermal cycling profile was used: 50°C for 30 minutes, 95°C for 15 minutes, then 35 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 minute. A final elongation step of 72°C for 10 minutes was used. Detection of conventional RT-PCR products was performed on a Qiaxcel (Qiagen, Inc) capillary electrophoresis system using a DNA screening cartridge and method AM420. Sequencing of open reading frame 5 (ORF5) was also attempted on at least one discrepant sample from these cases using forward primer 5'-AAG GTG GTA TTT GGC AAT GTG TC-3' and reverse primer 5'-GAG GTG ATG AAT TTC CAG GTT TCT A-3' and the qScript Custom One-Step RT-PCR Kit (Quanta Biosciences, Gaithersburg, Maryland). Polymerase chain reaction setup followed the manufacturer’s recommendations, with each primer included at 320 nM and the following cycling conditions: 48°C for 20 minutes, then 94°C for 3 minutes, followed by 45 cycles of 94°C for 30 seconds, 50°C for 50 seconds, and 68°C for 50 seconds. The final elongation step was 68°C for 7 minutes. Detection of PCR products was performed with the Qiaxcel instrument as described above. Polymerase chain reaction products of the correct size (1082 bp) were purified with ExoSAP-IT (Affymetrix/USB Products, Santa Clara, California) following the manufacturer’s recommendations, and were submitted to the Iowa State University DNA Facility for sequencing. Sequences were compiled using Lasergene software (DNAStar, Madison, Wisconsin).
Results
A total of 934 samples representing 423 clinical cases were tested with both VetMax (AB) and VetAlert (TC) PRRSV real-time RT-PCR reagents. Many cases consisted of more than one sample and in some cases, more than one specimen type. Case results were considered discrepant if at least one sample from a particular case was positive by one assay, with all samples from the case being negative by the other assay. From this testing, 27 cases (of 423) exhibited discrepant results. A subset of 19 of the discrepant cases were subjected to further testing by both assays, and 10 of these repeated as discrepant. Of those 10, six were lung samples, two were serum, and two were oral fluid. Table 1 summarizes sample types tested and results. The most common observations for those that did not repeat as discrepant were first, the original test was negative on pooled sera and positive on one or more individual samples from the pool when retested (n = 3), and second, the original Ct was lower than 36 cycles, which is considered weakly positive, and the original result did not repeat (n = 5). One sample had an original Ct of 28.15 only with the AB test, but upon retest was positive with both assays. Original and retest results of these nine cases are summarized in Table 2. Of the 10 repeating discrepant cases, nine were positive by the AB assay and negative with the TC reagents. The remaining discrepant case was positive by TC and negative by the AB assay.
Table 1: Summary of swine sample types and results from the comparison of two PRRSV real-time RT-PCR assays*
* Samples were submitted to the Iowa State University Veterinary Diagnostic Laboratory as routine diagnostic cases. Assays used were VetMax (Applied Biosystems, Foster City, California) and VetAlert (Tetracore, Rockville, Maryland). Cases included multiple samples and sometimes multiple sample types. † Discrepant cases exhibited a positive result on one or more samples with one assay and all negative results with the other assay. Repeating cases were those with the same qualitative results on the original test and retest. ‡ Mixture of undefined tissues submitted for testing. PRRSV = porcine reproductive and respiratory syndrome virus; RT-PCR = reverse transcriptase polymerase chain reaction; ORF = open reading frame; NA = not applicable; BAL = bronchoalveolar lavage fluid. |
Table 2: Values for cycle threshold (Ct) in swine cases for which original and retest qualitative results of two commercially available specific real-time RT-PCR assays for PRRSV differed*
* Assays were performed on clinical specimens submitted to the Iowa State University Veterinary Diagnostic Laboratory (Iowa State University, Ames, Iowa) with Ct defined as the cycle in the PCR reaction at which the fluorescent curve crossed the threshold when analyzed with the indicated settings; Ct > 40 considered negative. Retesting was performed on the original sample, or if the original sample was a serum pool, on individual samples from the pool. Assays used were VetMax (Applied Biosystems, Foster City, California) (AB) and VetAlert (Tetracore, Rockville, Maryland) (TC). † Individual samples from original pool. RT-PCR = reverse transcriptase polymerase chain reaction; PRRSV = porcine reproductive and respiratory syndrome virus; NA = not applicable (ie, original sample was not a pool). |
The 10 cases that repeated as discrepant were subjected to additional PRRSV testing by conventional RT-PCR and ORF5 sequencing attempts. The purpose for sequencing was twofold: first, to confirm the presence of PRRSV in the sample, and second, to determine relatedness of the viruses involved. Five of the cases positive by the AB test and negative by the TC test were confirmed PRRSV-positive by conventional RT-PCR. Two of the five also yielded complete or partial ORF5 sequences consistent with NA PRRSV (data not shown). The one case positive by TC and negative by AB was also confirmed as NA PRRSV-positive by conventional RT-PCR and sequencing. As an additional measure to check for contamination, these three sequences were compared to sequences of 44 samples from this study that had been determined at the request of the client in the diagnostic investigation, as well as the NA PRRSV used as positive control for the PCR testing. The dendrogram is shown in Figure 1. The sequence from the TC-positive, AB-negative sample (sequence 1) was 100% identical to one of the other sequences from this study. These two cases were submitted and tested 37 days apart. The one submitted earlier was a very weak positive sample (Ct 37.4) and was detected only by the TC assay, whereas the later submission was very strongly positive and detected by both assays (TC and AB Cts of 19.5 and 20.3, respectively). The sequences of the two samples positive by AB and negative by TC (sequences 2 and 3) were 84.2% homologous to each other and at most 96.8% homologous to any of the other sequences in the comparison. Confirmation by conventional RT-PCR or sequencing was not achieved for the other four discrepant cases. A total of 267 samples tested positive by both assays. For 154 of these (57.7%), the difference in Ct between the two assays was less than one cycle. In 50.6% of the 154 samples, the Ct was lower for the AB assay; the remaining 49.4% exhibited a lower Ct for the TC test.
Figure 1: Dendrogram of sequences obtained from samples yielding discrepant results as well as other samples in a study (described in Table 1) comparing two porcine reproductive and respiratory syndrome virus (PRRSV) real-time reverse transcriptase polymerase chain reaction (RT-PCR) assays. Assays used were VetMax (Applied Biosystems, Foster City, California) (AB) and VetAlert (Tetracore, Rockville, Maryland) (TC). Arrows indicate the one sample (serum) positive by TC and negative by AB (1) and the two samples (serum and oral fluid, respectively) positive by AB and negative by TC (2,3). Positive control (pos control) was the North American PRRSV used as positive control in the RT-PCR assays. 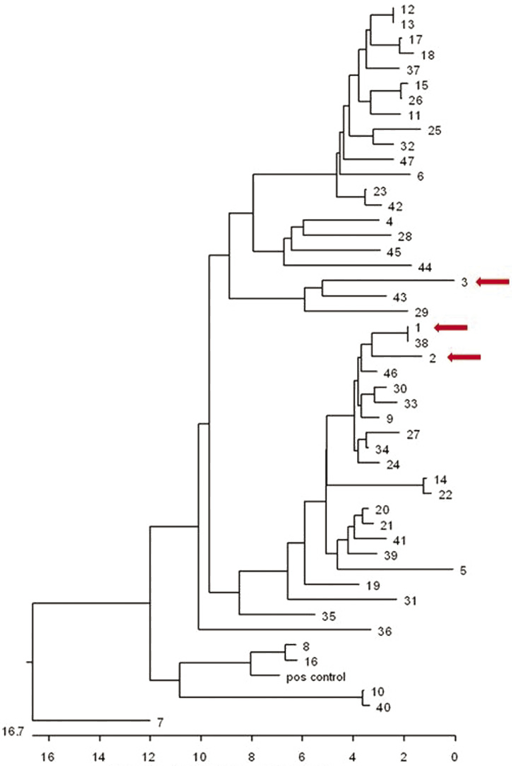 Nucleotide substitution per 100 residues |
Discussion
In this study, a comparison of two commercially available PRRSV-specific real-time RT-PCR assays was performed. The data were evaluated on a clinical-case basis, rather than on a per-sample basis that assumes that discrepancies between assays for individual samples within a positive case are due to differences in sensitivity rather than specificity. This is based on the assumption that the PRRS viruses in the samples within a case have a very high degree of similarity, which may be a false assumption, but many of the discrepant samples were weakly positive and could not be sequenced to confirm the PRRSV strain. The majority of positive samples were detected by both assays, with similar Cts. This indicates that the settings that were used for the analysis gave similar results across a broad range of samples, and did not give either assay an unfair advantage regarding sensitivity. The discrepant results were observed in oral fluid, lung, and serum samples, which was not unexpected since they comprised the three specimen types of greatest number in the study. Discrepant results that did not repeat were most often from weak positive samples or pooled samples. We were unable to explain the lack of repeatability for the one remaining sample, but this most likely was due to either a pipetting or sampling error during the original testing. Comparison of ORF5 sequences obtained in this study revealed that the two cases detected by AB and missed by TC were not closely related. The sequence of the one case detected by TC and missed by AB was identical to the sequence derived from one other sample in the study. These two cases were submitted and tested more than 1 month apart, and test results of the two assays were inconsistent between the two submissions, so it is highly unlikely that the discrepant results from this sample were due to contamination. This study underscores the risk in evaluating animal or herd status by testing pooled samples. In three cases, a pooled sample was determined to be negative, but when the individual samples from that pool were tested, at least one sample was positive. Many submitters prefer to test pools rather than individual specimens, but caution must be taken in interpreting and utilizing this data to make management decisions. See Rovira et al8 for a more thorough discussion of this topic.
Both assays used in the current study have multiple primers and probes to enhance their ability to detect contemporary PRRS viruses. The previous version of the AB test had missed or weakly detected numerous PRRSV-positive cases that the TC assay detected (observed and confirmed in Iowa State University Veterinary Diagnostic Laboratory, 2010). The assay was updated using information and samples submitted to AB by our laboratory and other users of the assay (personal communication, AB company personnel, 2010). The performance of the updated AB assay was much improved over the former version. However, as evidenced by this study, false-negative results were still observed with both sets of reagents, indicating the likelihood of mismatches between primer and probe sequences and the target region, which can affect both sensitivity and specificity of a PCR reaction. Involvement of the assay manufacturer is critical to ensure that the reagents are updated to detect the currently circulating viruses. To the extent possible, the samples from this study that yielded discrepant results were submitted to the appropriate company for use in further evaluating their assay. Likewise, client feedback concerning unexpected results is vital for initiating subsequent testing on the samples in question, as well as for alerting laboratory staff to potential problems with the assay and the need for updating the reagents.
Implications
• Client communication concerning unexpected results is critical for proper follow-up PCR testing, as well as for alerting the laboratory to the possible requirement for assay improvement.
• Pooling samples for testing may result in false-negative results, and care must be taken when making decisions on testing regimens and utilizing results.
• RNA viruses such as PRRSV mutate, resulting in genetic variability and possible failure of RT-PCR tests due to mismatches in assay target regions. Involvement of assay manufacturers is vital for ensuring reagents are updated as necessary.
References
1. Ahl R, Pensaert M, Robertson IB, Terpestra C, van der Sande W, Walker KJ, White E, Meredith M. Porcine reproductive and respiratory syndrome (PRRS or blue-eared pig disease). Vet Rec. 1992;130:87–89.
2. Chang CC, Yoon KJ, Zimmerman JJ, Harmon KM, Dixon PM, Dvorak CMT, Murtaugh MP. Evolution of porcine reproductive and respiratory syndrome virus during sequential passages in pigs. J Virol. 2000;76:4750–4763.
3. Klungthong C, Chinnawirotpisan P, Hussem K, Phonpakobsin T, Manasatienkij W, Ajariyakhajorn C, Rungrojcharoenkit K, Gibbons RV, Jarman RG. The impact of primer and probe-template mismatches on the sensitivity of pandemic influenza A/H1N1/2009 virus detection by real-time RT-PCR. J Clin Virol. 2010;48:91–95.
4. Ratcliff RM, Chang G, Kok T, Sloots TP. Molecular diagnosis of medical viruses. Curr Issues Mol Biol. 2007;9:87–102.
5. Papin JF, Vahrson W, Dittmer DP. SYBR green-based real-time quantitative PCR assay for detection of West Nile virus circumvents false-negative results due to strain variability. J Clin Microbiol. 2004;42:1511–1518.
6. Chittick WA, Stensland WR, Prickett JR, Strait EL, Harmon K, Yoon KJ, Wang C, Zimmerman JJ. Comparison of RNA extraction and real-time reverse transcription polymerase chain reaction methods for the detection of porcine reproductive and respiratory syndrome virus in porcine oral fluid specimens. J Vet Diagn Invest. 2011;23:248–253.
7. Christopher-Hennings J, Nelson EA, Nelson JK, Hines RJ, Swenson SL, Hill HT, Zimmerman JJ, Katz JB, Yaeger MJ, Chase CCL, Benfield DA. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J Clin Microbiol.1995;33:1730–1734.
8. Rovira A, Clement T, Christopher-Hennings J, Thompson B, Engle M, Reicks D, Munoz-Zanzi C. Evaluation of the sensitivity of reverse-transcription polymerase chain reaction to detect porcine reproductive and respiratory syndrome virus on individual and pooled samples from boars. J Vet Diagn Invest. 2007;19:502–509.
