| Original research | Peer reviewed |
Cite as: Cuong NV, Carrique-Mas J, Thu HTV, et al. Serological and virological surveillance for porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, and influenza A viruses among smallholder swine farms of the Mekong Delta, Vietnam. J Swine Health Prod. 2014;22(5):224–231.
Also available as a PDF.
SummaryObjectives: To evaluate the feasibility and utility of oral-fluids collection for surveillance of porcine viruses in the Mekong Delta, Vietnam, and to establish baseline serological and virological prevalence estimates for porcine reproductive and respiratory syndrome virus (PRRSV), porcine circovirus type 2 (PCV2), and influenza A virus (IAV) among smallholder farms. Materials and methods: Paired serum and oral-fluids samples from 68 farms (sows, boars, weaners, and growers) were tested during 2011 by reverse transcriptase polymerase chain reaction and enzyme-linked immunosorbent assay for PRRSV, PCV2, and IAV. Results: Low numbers of PRRSV-positive and IAV-positive pigs were detected (1.6% PRRSV viremic, two of 124; 0.8% IAV in oral fluids, one of 124). However, PCV2 detection rates were high in both serum and oral fluids (54.8% and 61.3%, respectively). Overall proportions of pigs seropositive for IAV and PRRSV were 37.9% and 33.9%, respectively. Proportions of pigs seropositive for PRRSV were 48.6% (17 of 35) and 12.1% (four of 33) on vaccinated and unvaccinated farms, respectively. Oral fluids and serum samples yielded comparable prevalence estimates for molecular detection of PCV2, and detected one sample PCR-positive for hemagglutinin of influenza A/H1N1/pdm09. There was no evidence of PRRSV shedding in oral fluids. Implications: Antibody prevalence estimates based on testing oral fluids may provide an acceptable and useful surrogate for testing serum in future field studies if optimized assays are employed. | ResumenObjetivos: Evaluar la viabilidad y utilidad de la recolección de fluidos orales para la vigilancia de virus porcinos en Mekong Delta, Vietnam, y para establecer valores base de la prevalencia serológica y virológica para el virus del síndrome reproductivo y respiratorio porcino (PRRSV por sus siglas en inglés), circovirus porcino tipo 2 (PCV2 por sus siglas en inglés), y el virus de la influenza A (IAV por sus siglas en inglés) en granjas de pequeños productores. Materiales y métodos: Se analizaron muestras de fluidos orales y sueros pareados de 68 granjas (machos, hembras, lechones de destete, y crecimiento) durante 2011 por medio de la prueba de reacción en cadena de polimerasa de transcriptasa reversa y la prueba de inmunoabsorción enzimática para PRRSV, PCV2, e IAV. Resultados: Se detectaron bajos números de cerdos positivos al PRRSV y positivos al IAV (1.6% virémicos al PRRSV, dos de 124; 0.8% IAV en fluidos orales, uno de 124). Sin embargo, los índices de detección de PCV2 fueron altos en sueros y fluidos orales (54.8% y 61.3%, respectivamente). En general, las proporciones de cerdos seropositivos al IAV y PRRSV fueron 37.9% y 33.9%, respectivamente. Las proporciones de cerdos seropositivos al PRRSV fueron 48.6% (17 de 35) y 12.1% (cuatro de 33) en granjas vacunadas y no vacunadas, respectivamente. Las muestras de suero y fluidos orales arrojaron valores de prevalencia comparables a la detección molecular de PCV2, y detectaron una muestra positiva al PCR para la hemaglutinina de influenza A/H1N1/pdm09. No hubo evidencia de excreción de PRRSV en fluidos orales. Implicaciones: Los cálculos de prevalencia de anticuerpos basados en pruebas de fluidos orales pueden ofrecer un sustituto aceptable y útil para probar suero en futuros estudios de campo si se emplean pruebas optimizadas. | ResuméObjectifs: Évaluer la faisabilité et l’utilité de la collecte de fluides oraux pour la surveillance de virus porcins dans le Delta du Mékong, Vietnam, et établir les estimés des prévalences sérologique et virologique de base pour le virus du syndrome reproducteur et respiratoire porcin (PRRSV), le circovirus porcin de type 2 (PCV2), et le virus de l’influenza A (IAV) dans des fermes de petite taille. Matériels et méthodes: Des échantillons pairés de sérum et des échantillons de fluides oraux provenant de 68 fermes (truies, verrats, porcs sevrés, porc en engraissement) ont été testés durant l’année 2011 par réaction d’amplification en chaine par la polymérase à l’aide de la transcriptase réverse et par épreuve immunoenzymatique (ELISA) pour PRRSV, PCV2, et IAV. Résultats: Des nombres peu élevés de porcs positifs pour PRRSV et IAV furent détectés (1,6% PRRSV virémiques, 2 sur 124; 0,8% pour IAV dans les fluides oraux, 1 sur 124). Toutefois, les taux de détection de PCV2 étaient élevés autant dans les échantillons de sérum que de fluides oraux (54,8% et 61,3%, respectivement). De manière générale, les taux de porcs séropositifs pour IAV et PRRSV étaient de 37,9% et 33,9%, respectivement. Les taux de porcs séropositifs pour le PRRSV étaient de 48,6% (17 sur 35) et 12,1% (4 sur 33) pour les fermes pratiquant et ne pratiquant pas la vaccination, respectivement. Les échantillons de fluides oraux et de sérum ont donné des résultats d’estimé de prévalence comparables pour la détection moléculaire de PCV2, et ont permis de détecter un échantillon positif par PCR pour l’hémagglutinine du virus influenza A/H1N1/pdm09. Il n’y avait aucune évidence d’excrétion de PRRSV dans les fluides oraux. Implications: Les estimés de prévalence des anticorps basés sur les épreuves effectuées sur les fluides oraux peuvent être des alternatives acceptables et utiles aux tests effectués sur du sérum si des épreuves optimisées sont utilisées. |
Keywords: swine, oral fluids, influenza, porcine reproductive and respiratory syndrome virus, PRRSV, Vietnam
Search the AASV web site
for pages with similar keywords.
Received: October 28, 2013
Accepted: February 27, 2014
Pork production is critically important to the national economy and food security of Vietnam, and despite major animal-disease outbreaks, the swine industry of Vietnam has achieved remarkably sustained growth in production and profitability over the last 30 to 40 years. Between 2000 and 2010, the total volume of pork production in Vietnam increased 114%.1 These increases were due to growth in pig stocks (approximately 3.6% annual growth in swine population between 2000 and 2010, from 23.0 to 49.3 million head), as well as increased efficiencies in production.2 Animal-health issues facing the industry include fatal epizootics of porcine viruses, endemic circulation of several notifiable diseases (eg, foot-and-mouth disease, classical swine fever), and additional pathogens that reduce efficiency and profitability, some of which may have zoonotic implications for human health (eg, influenza A viruses, Streptococcus suis, Salmonella serovars, Trichinella species, cysticercosis).2-5
Major epizootics of porcine high fever disease (PHFD) caused devastating losses to the Vietnamese swine sector in 2007-2010, impacting 53 of 63 provinces and resulting in more than 1,100,000 pigs destroyed in 2010 alone.6 The principle agent suspected in these outbreaks was porcine reproductive and respiratory syndrome virus (PRRSV). Although PRRSV was clearly a major driver of the explosive outbreaks, experimental studies using a Vietnamese isolate of PRRSV failed to reproduce the severe clinical syndromes seen in the field,7 suggesting possible co-infections or other co-factors contributing to the highly pathogenic phenotype. Among the agents suspected of involvement were porcine circovirus type 2 (PCV2), classical swine fever virus, and various bacterial agents (eg, Pasteurella multocida, S suis, Mycoplasma hyopneumoniae, Haemophilus parasuis, and Actinobacillus pleuropneumoniae).8 During the PHFD outbreaks of 2007-2010, PRRSV and PCV2 were detected in 80% and 90% of swine cases, respectively, submitted to the National Center for Veterinary Diagnostics, Hanoi, Vietnam.9 During 2009-2011, approximately 60% of PHFD outbreaks were confirmed positive for PRRSV, while the remaining 40% were negative for PRRSV but positive for PCV2 or other co-infecting agents.
Influenza A viruses (IAVs) circulating in pigs are of particular concern for the Mekong Delta region due to the endemic circulation of highly pathogenic avian influenza (HPAI) within domestic poultry populations,10 the frequency of mixed rearing of pigs and poultry in backyard farming operations,11 and the potential role of swine in the emergence of avian-swine-human reassortant viruses.12 Data from the Mekong Delta suggest that all three major lineages of IAVs in swine (classical swine H1N1, Eurasian avian-like swine H1N1, and North American triple reassortant viruses) co-circulate.13 Although neither HPAI H5N1 nor low pathogenic avian influenza viruses have been isolated yet from pigs in Vietnam, a novel human-swine reassortant H3N2 was detected in Vietnamese pigs in 2010.5 Studies of IAV in Vietnamese pigs have shown significant geographic variability in seroprevalence,14 from very low levels of circulation (3.1% positive) in semi-commercial farms in a remote northern province15 to 65% seropositive in intensive farms of the Red River Delta.16
Despite the critical imperatives for improved surveillance of swine diseases, the network for animal-disease reporting lacks resources, and veterinary laboratory diagnostics are rarely available, hence few samples are submitted for confirmatory analyses. The lack of baseline prevalence data is due in part to the logistical and technical challenges of sampling animals from small backyard operations; among Vietnamese households raising pigs, approximately 91% have fewer than 10 pigs, and only 6% have more than 30 pigs.11 Veterinary extension services are limited, and farmers are generally reluctant to restrain animals for collection of blood or nasal swabs. Oral fluids are a diagnostic specimen for detection of many human and veterinary pathogens, and are of increasing interest for routine surveillance activities.17-19 To the authors’ knowledge, oral-fluids-based surveillance has not been evaluated within the context of smallholder farming systems in Vietnam. We hypothesized that oral fluids would present a viable alternative to serum samples for routine surveillance and would assist in overcoming farmer reluctance to sampling, particularly of young piglets. We therefore evaluated the performance of individual and pen-based oral-fluids diagnostics for three of the most important porcine respiratory viruses, PRRSV, PCV2, and IAV, in a province of the Mekong Delta that had previously experienced outbreaks of PHFD.
Materials and methods
The survey did not require ethical review because the activities comprised part of periodic routine postvaccination monitoring, did not involve animal experimentation, and were implemented by the relevant animal-health authorities of the province.
The survey was implemented by the Subdepartment of Animal Health (SDAH) of Can Tho province within the context of periodic routine postvaccination monitoring. The survey was carried out in September 2011 in the Can Tho province of Southern Vietnam, located between latitudes 9°55'08″ and 10°19'38″ north and longitudes 105°13'38″ and 105°50'35″ east. With an area of 1409 km2, the province is home to approximately 1.2 million people and 5343 pig farms with approximately 126,000 pigs (2011 agricultural census). The province has a total of nine districts and 85 communes. Farms were selected at random (using coin toss and census lists of registered farms) from 21 communes within the eight districts that had a history of confirmed porcine reproductive and respiratory syndrome (PRRS) in 2010 as determined by the SDAH in Can Tho. The number of farms sampled was proportional to the number of farms in the study communes. The study aimed to collect up to six oral-fluids samples and up to 12 blood samples per farm. Farmer consent was obtained with financial compensation, as per standard SDAH practice. Samples were collected from individually confined and group-penned animals. For individually confined animals (sows, boars), one oral-fluids sample was collected per animal. For group-penned pigs (weaners and growers up to 50 weeks old), pen-based oral fluids were collected. Blood samples were collected only from pigs that contributed to oral-fluids collections (ie, were observed to actively chew on ropes).
Animal sampling method
The protocol for oral-fluids collection was first tested in a pilot study on a local farm. We selected locally produced, 100% cotton, 2-cm diameter woven rope, which was cut into 100-cm sections and unraveled for approximately 10 cm at one end. Ropes were tied to the railings of each pen, and pigs were allowed to chew for 20 minutes under continuous observation. The wet portion of the rope was inserted into a 1-litre re-sealable plastic bag and hand-wrung to extract the fluids; 2 mL was transferred to a cryovial and immediately flash frozen in a liquid nitrogen vapor-cooled Dewar dry shipper (-140°C) to ensure optimal conditions for subsequent virological testing. After completion of oral-fluids collections, pigs that had actively chewed were restrained by rope, and 6 to 8 mL of whole blood was collected by jugular venipuncture. Serum separation, aliquoting, and transfer to temporary storage at -20°C were performed within approximately 6 hours of collection. All sample collections were transferred to -80°C within 1 week of collection. Sample identification enabled linkage between serum and oral-fluids samples.
Sample processing
Serum samples were analyzed both individually and pooled for detection of viral pathogens. Pools were prepared by mixing 100 µL of each sample to reflect the same aggregates as the pen-based oral fluids. Nucleic acids (NA) were extracted from sera and oral fluids using 200 µL and the MagNA Pure 96 Viral NA small volume kit (Roche, Basel, Switzerland) and an automated extractor (Roche). Presence of polymerase chain reaction (PCR) inhibitors and NA quality control were assessed by spiking samples with an RNA internal extraction control (equine arterivirus) prior to extraction.20 The total RNA recovered (60 µL in nuclease-free water) was stored at -80°C until use. Real-time reverse transcriptase PCR (RT-PCR) was performed using primers and probes described for PRRSV21 and matrix gene of IAV,22 using SuperScriptIII Platinum One-Step Quantitative kits (Invitrogen, Carlsbad, California) performed in a 25-µL reaction mix on a Chromo4 real-time PCR machine (Biorad, Hercules, California). Molecular screening for influenza was limited to oral-fluids samples, because IAV is not known to cause viremia in swine. Oral fluids positive for IAV by matrix gene PCR were further tested using primer pairs for a swine-specific influenza A nucleoprotein (NP) gene and hemagglutinin subtyping primers for A/H1N1/pdm09, human H3, and avian H5 lineages (current US Centers for Disease Control subtyping primers). Additional testing was subsequently performed using pan-hemagglutinin23 and pan-neuraminidase24 primers, a 2× PCR enzyme mix as described for oral-fluids optimization,25 and products detected by conventional gel electrophoresis. Virus isolation for IAV-positive samples was attempted in embryonated chicken eggs (three eggs per sample) and concurrently for three serial passages in Madin-Darby canine kidney (MDCK) cells.22 For PCV2 detection, amplifications were performed using primers and probes26 that had been used in previous studies in southern Vietnam.27 The real-time PCV2 PCR was performed in a 25-µL format using TaqMan Universal PCR Master Mix (Applied Biosystems, Carlsbad, California) and Lightcycler480 (Roche).
PRRS virus antibody detection was performed on serum and oral fluids using the HerdChek PRRS X3 (Idexx Laboratories, Westbrook, Maine), designed to detect Chinese, European, and North American lineages of PRRSV. Serum samples were processed according to manufacturer’s instructions, whereas oral-fluids processing was modified by decreasing the dilution (1:2 instead of 1:20) and using larger volumes (250 versus 100 µL) and longer incubation (16 hours versus 1 hour).28 Influenza antibody detection was performed using Influenza A Antibody Test (Idexx Laboratories, Westbrook, Maine), and both serum samples and oral fluids were processed identically following the manufacturer’s instructions.
Statistical analyses
The interquartile range (IQR) of pigs per farm was calculated. Diagnostic yields (numbers of positives) of oral-fluids versus serum samples (representing the same sampled animals) were compared using McNemar’s chi-square test; the kappa test was used to measure the level of agreement among tests.29 The following benchmarks were used for interpretation of kappa test results: 0 to < 0.01 = poor; 0.01 to 0.20 = slight; 0.21 to 0.40 = fair; 0.41 to 0.60 = moderate; 0.61 to 0.80 = substantial; 0.81 to 1 = almost perfect. For PRRSV antibody detection, results from individual serum samples (N = 313), pooled serum samples (N = 84), individual oral-fluids samples (N = 40), and pooled oral-fluids samples (N = 84) were stratified by PRRSV vaccination status and history of disease compatible with PRRS on the farm (abortion in sows and respiratory signs in weaners and growers). All comparisons were made using the chi-square test. Analyses were carried out using R software within the EpiR package (http://www.r-project.org/). Comparisons were considered significant at P < .05.
Results
Farm characteristics and sample collection
A total of 68 farms from 21 communes in eight districts were sampled. Of the 68 farms surveyed, 25 (36.8%) were small-scale household farms with three to five sows; 30 (44.1%) were medium size (six to 20 sows); six (8.8%) were larger commercial units (> 20 sows); and seven (10.3%) raised only growers or finishers. The median total number of pigs (all ages) per farm was 24.5 (IQR 16.0 to 75.4), which is representative of the median farm size in the province. Twenty-three farms (33.8%) reported a history consistent with PRRSV infection (abortion in sows or respiratory signs in piglets), as determined by SDAH, and 36 (52.9%) reported vaccination against PRRSV over the past 12 months. Other diseases for which vaccination was carried out included classical swine fever (79.3% of farms); pasteurellosis (69.8% of farms); salmonellosis (67.2% of farms); and foot-and-mouth disease (56.7% of farms).
A total of 124 oral-fluids samples were collected. These corresponded to 40 animals individually penned (gilts, sows, and boars) and 84 animals in pens with ≥ 8 individuals (mostly weaners, growers, and some gilts, range eight to 15) (n = 84 groups). Upon initial exposure to the ropes, most pigs engaged in active chewing. One pen was sampled from 35 farms (51.5%); two pens were sampled from 18 farms (26.5%); three pens were sampled from 10 farms (14.7%); and four to six pens were sampled from five farms (7.4%). Blood was collected from a total of 313 pigs, which were the same animals (individuals or groups) observed chewing the ropes and from which oral fluids were collected (40 from individually penned animals and 273 from 84 pens with eight to 15 animals each).
Virus detection by PCR in oral fluids and serum
Summary results for the tests performed in matched oral fluids and serum are presented in Table 1. Porcine circovirus type 2 DNA was detected in 54.8% and 61.3% of serum and oral-fluids samples, respectively, indicating that assay sensitivity did not differ significantly by specimen type (Table 2). Results of paired comparisons of oral-fluids and serum samples from individual pigs were more concordant (fair agreement) than those obtained with pooled oral-fluids and serum samples (Table 2). Estimates of overall farm-level, oral-fluids antibody prevalence for IAV and PRRSV did not differ (14.7%, 10 of 68 in each case); however, estimates for pathogen prevalence that were based on serum antibody were significantly different (IAV 23.5%, 16 of 68) and (PRRSV 8.8%, six of 68) (P < .5).
Table 1: Results of virological (PCR) and antibody prevalence testing (ELISA*) for PCV2, IAV, and PRRSV in oral fluids and serum collected from pigs on 68 farms in Can Tho province, Vietnam, during 2011†
| N | Virus testing (PCR) | Antibody testing | ||||
|---|---|---|---|---|---|---|
| PCV2 | IAV | PRRSV | IAV | PRRSV | ||
| No. pos (%) | No. pos (%) | No. pos (%) | No. pos (%) | No. pos (%) | ||
| Oral-fluids samples | ||||||
| Individual | 40 | 23 (57.5) | 0 (0) | 0 (0) | 12 (30.0) | 9 (22.5) |
| Pen-based | 84 | 53 (63.1) | 1 (1.2) | 0 (0) | 24 (28.6) | 22 (26.2) |
| Total samples | 124 | 76 (61.3) | 1 (0.8) | 0 (0) | 36 (29.0) | 31 (25.0) |
| Serum samples | ||||||
| Individual | 40 | 18 (45.0) | ND | 1 (2.5) | 22 (55.0) | 23 (57.5) |
| Pen (pooled) | 84 | 54 (64.3) | ND | 1 (1.2) | 25 (29.8) | 19 (22.6) |
| Total samples | 124 | 68 (54.8) | ND | 2 (1.6) | 47 (37.9) | 42 (33.9) |
* HerdChek PRRS X3 ELISA (Idexx Laboratories, Westbrook, Maine). Serum samples were processed according to manufacturer’s instructions. For oral fluids, dilution was 1:2 (versus 1:20), volume was 250 µL (versus 100 µL), and incubation time was16 hours (versus 1 hour).
† For individually confined animals (gilts, sows, and boars), one oral-fluids sample was collected per animal. For group-penned pigs (weaners and growers up to 50 weeks old) pen-based oral fluids were collected (eight to 15 animals/pen). Blood samples were collected only from pigs that contributed to oral-fluids collections, ie, were observed actively chewing ropes.
PCR = polymerase chain reaction; ELISA = enzyme-linked immunosorbent assay; PCV2 = porcine circovirus type 2; IAV = influenza A virus; PRRSV = porcine reproductive and respiratory syndrome virus; pos = positive; ND = not done.
Table 2: Detection by PCR of PCV2 viral DNA from 124 oral fluids (OF) and pooled serum (S) samples from pigs surveyed in this study*
| N | OF(+) PCV2 (%) | S(+) PCV2 (%) | OF(+) S(+) | OF(-) S(-) | OF(+) S(-) | OF(-) S(+) | McNemar | Kappa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| χ2 | P | Kappa | Level of agreement† | ||||||||
| Individual samples | 40 | 23 (57.5) | 22 (55.0) | 15 | 10 | 8 | 7 | 0 | 1 | 0.24 | Fair |
| Pooled samples | 84 | 53 (63.1) | 46 (54.8) | 29 | 14 | 24 | 17 | 0.88 | .35 | -0.001 | Poor |
| All samples | 124 | 76 (61.3) | 68 (54.8) | 44 | 24 | 32 | 24 | 0.87 | .35 | 0.08 | Slight |
* Study described in Table 1.
† For kappa test results: 0 to < 0.01 = poor; 0.01 to 0.20 = slight; 0.21 to 0.40 = fair; 0.41 to 0.60 = moderate; 0.61 to 0.80 = substantial; 0.81 to 1 = almost perfect. Comparisons were considered significant at P < .05.
PCR = polymerase chain reaction; PCV2 = porcine circovirus type 2; (+) = positive; (-) = negative.
Influenza viral RNA was identified by matrix gene detection in one oral-fluids sample from penned growers (0.8%, one of 124). This sample was confirmed positive using NP primers designed to detect all contemporary swine influenza lineages, and primers for the HA of A/H1N1/pdm09. Virus isolation attempts in eggs and MDCK cells were unsuccessful and depleted the original sample volume. Subsequent attempts to generate amplicons from stored RNA extractions using pan-HA and pan-NA primers did not yield quality sequence reads, and above-threshold cycle threshold (Ct) values for internal RNA controls suggested poor sample quality.
All 124 oral-fluids samples tested negative for PRRSV by RT-PCR, whereas two serum samples tested positive (one pooled sample from a pen of growers with Ct value = 30 and one individually tested sow serum sample with Ct value = 24). The farm with a single pen of PRRSV-positive growers was a relatively large operation (100 sows, total > 400 pigs) and reported prior use of PRRSV vaccine, although the farmer could not specify the manufacturer. These growers also tested positive for PRRSV antibody in the corresponding pooled oral-fluids sample, but not in the pooled serum sample. The PRRSV PCR-positive sow was from a household with two sows and six piglets, and the farmer reported no PRRSV vaccination. The sow tested negative by ELISA for PRRSV antibody in both oral-fluids and serum samples.
Antibody detection by ELISA in oral fluids and serum
Antibody detection for IAV and PRRSV in oral-fluids versus serum samples is presented in Table 3. Overall, antibody testing for IAV was more sensitive for serum than for oral-fluids samples, and there was moderate agreement between the sample types. In individually tested pigs, there was a larger differential in antibody prevalence between serum and oral-fluids samples. For pooled samples, sensitivities of the sample types did not differ for IAV antibody detection. This pattern was similar for PRRSV antibody detection; prevalence of PRRSV antibody detection was greater in serum samples than in oral-fluids samples, and antibody prevalence was greater when individual samples were tested rather than pools. There was fair to moderate agreement between oral-fluids samples and serum samples in all cases.
Table 3: Detection of IAV and PRRSV antibodies in porcine oral fluids (OF) and serum (S) tested by commercial ELISA (HerdChek PRRS X3, Idexx Laboratories, Westbrook, Maine)*
| Antibody test | Sample type | No. | OF(+) IAV antibody (%) | S(+) IAV antibody (%) | OF(+) S(+) | OF(-) S(-) | OF(+) S(-) | OF(-) S(+) | McNemar | Kappa | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| χ2 | P | Kappa | Level of agreement† | |||||||||
| IAV | Individual samples | 40 | 30.0 | 55.0 | 12 | 18 | 0 | 10 | 8.10 | <.01 | 0.51 | Moderate |
| Pooled samples | 84 | 28.6 | 29.8 | 16 | 51 | 8 | 9 | 0 | 1 | 0.51 | Moderate | |
| All samples | 124 | 29.0 | 37.9 | 28 | 69 | 8 | 19 | 3.70 | .05 | 0.51 | Moderate | |
| PRRSV | Individual samples | 40 | 22.5 | 57.5 | 8 | 16 | 1 | 15 | 10.56 | <.001 | 0.26 | Fair |
| Pooled samples | 84 | 22.6 | 27.4 | 10 | 52 | 9 | 13 | 0.49 | .52 | 0.30 | Fair | |
| All samples | 124 | 22.6 | 37.1 | 18 | 68 | 10 | 28 | 7.60 | .01 | 0.29 | Fair | |
* Pigs and sampling described in Table 1.
† For kappa test results: 0 to < 0.01 = poor; 0.01 to 0.20 = slight; 0.21 to 0.40 = fair; 0.41 to 0.60 = moderate; 0.61 to 0.80 = substantial; 0.81 to 1 = almost perfect. Comparisons were considered significant at P < .05.
IAV = influenza A virus; PRRSV = porcine reproductive and respiratory syndrome virus; ELISA = enzyme-linked immunosorbent assay; (+) = positive; (-) = negative.
PRRSV ELISA testing results by age, vaccination status, and history of disease on farms
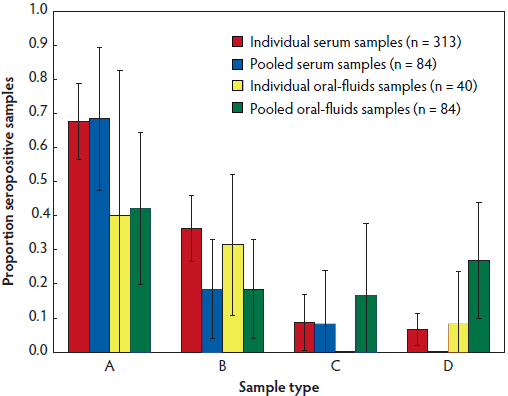
Comprehensive ELISA testing of the 313 individual serum samples yielded an overall PRRSV seropositivity of 29.1% (24.0% to 34.1%). Older pigs had a greater probability of testing seropositive (Figure 1). Overall PRRSV seropositivity in vaccinated versus unvaccinated farms was 48.6% (17 of 35) and 12.1% (four of 33), respectively. The highest rates of PRRSV seropositivity were found among farms that had vaccinated for PRRSV and had a history of PRRSV disease (67.6%; 95% CI, 68.4%-40.0%), and the lowest for farms with no history of PRRSV or vaccination (6.7%; 95% CI, 1.9%-11.4%; P < .001, χ2). No statistical differences in rate of seropositivity were observed between samples from unvaccinated farms with and without history of PRRSV disease (8.7% versus 6.7%; P = .99, χ2) (Figure 2). Pooled oral-fluids samples from unvaccinated farms with no history of PRRSV had an unusually high prevalence of seropositivity (26.9%; 95% CI, 9.9%-44.0%).
Figure 1: Porcine reproductive and respiratory syndrome virus (PRRSV) antibody detection by ELISA was conducted on individual pig serum samples collected from small farms in Can Tho province, Vietnam, in 2011, as part of routine post-PRRSV vaccination monitoring. Serum samples were stratified by age in months (N = 313). Samples were tested using the HerdChek PRRS 3X ELISA (Idexx Laboratories, Westbrook, Maine). The trend of increasing proportion of PRRSV seropositivity with age was significant (P < .05: chi-square).
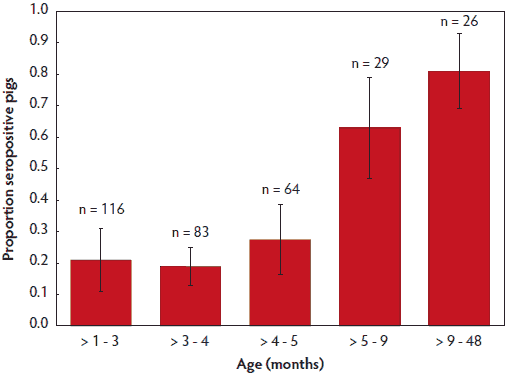
Figure 2: Prevalence of detection of antibody against porcine reproductive and respiratory syndrome virus (PRRSV) in individual animal serum samples (n = 313), pooled serum samples (n = 84), individual oral-fluids samples (n = 40), and pooled oral-fluids samples (n = 84) from pigs in small farms in Can Tho province, Vietnam, as described in Table 1. A = samples from farms with both history of PRRSV and PRRSV vaccination; B = samples from farms with PRRSV vaccination and no history of PRRSV; C = samples from farms with history of PRRSV and no PRRSV vaccination; D = samples from farms with neither history of PRRSV nor PRRSV vaccination. Samples were tested using the HerdChek PRRS 3X ELISA (Idexx Laboratories, Westbrook, Maine). Differences between A and D were significant (P < .001; chi-square).
Discussion
Our virological and serological analyses confirm endemic co-circulation of PRRSV, PCV2, and IAV within one southern province of Vietnam. We report low levels of PRRSV viremia and IAV shedding in oral fluids, and high levels of both viremia and shedding in oral fluids for PCV2. Antibody detection was more sensitive in serum samples than in oral fluids for both IAV and PRRSV, and there was fair to moderate concordance between the two sample types. Regarding diagnostic efficacy for molecular screening, oral-fluids samples yielded promising results for PCV2 and IAV, but no detections of PRRSV. Detection of PCV2 viral DNA was comparable in oral-fluids and serum samples. In the older pigs, PCV2 was detected significantly more often in oral-fluids samples than in serum samples, indicating prolonged shedding of PCV2 from the respiratory tracts of mature pigs (in contrast to resolution of systemic viremia).
Since our study implementation and sample processing, a number of published investigations have highlighted the need to specifically tailor diagnostic assays for the oral-fluid matrix30-32 and have thoroughly evaluated the use of oral fluids for monitoring herd health. Panyasing et al31 document modifications to an influenza blocking NP ELISA similar to those described by Kittawornrat et al33 for PRRS ELISA, with reportedly better results; these modifications were not used in the present study. Our failure to find high concordance between assay results for oral fluids and serum for all three pathogens (in particular for PRRSV) are not consistent with the recent reports and may reflect important technical deficiencies in our sample processing. Our results may also reflect inherent variability or bias when evaluating diagnostic protocols using relatively small sample sizes or populations with overall low prevalence of viral shedding.
The oral-fluids screening for IAV yielded one positive (one of 124; 0.8%), and subtyping by PCR confirmed that the sample was positive for hemagglutinin of A/H1N1/pdm09. Because we were unable to confirm the partial HA or NA amplicons by sequencing and did not sequence internal gene fragments, it remains unclear whether the detected virus was similar to pH1N1 currently circulating in people, or was an independent lineage or mixed virus. We anticipate further reports from government swine-surveillance activities that will clarify the complex situation of co-circulating reassortant subtypes in the region. The fact that IAV virus isolation from oral fluids was not successful suggests the presence of virus-inactivating factors within saliva (such as IAV antibodies), dilution effects in saliva, or sample degradation that impaired infectivity but did not entirely degrade RNA. Current swine surveillance programs continue to focus exclusively on use of nasal-swab specimens, and it remains to be seen whether optimization protocols for oral-fluids virus isolations will be accepted in the Vietnam context.
Conventional individual testing of serum samples by PRRSV ELISA revealed the expected age-dependent increase in PRRSV seropositivity, as well as a significant relationship between seropositivity, PRRS vaccination status, and history of PRRSV disease on farms. The observed PRRSV seropositivity in approximately 12% of unvaccinated farms that did not report PRRSV disease might reflect asymptomatic seroconversion to wild-type field virus, inaccuracy in reporting PRRSV vaccination status, secondary transmission of live attenuated vaccine virus, or all three. The JAX-1 vaccine (based on an attenuated virus of the highly pathogenic Chinese lineage) has been licensed for use in Vietnam since 2008, but was not used in Can Tho province during the time of the survey collections in 2011. The vaccines used on the survey farms at that time were commercial vaccines from Singapore, Germany, and Spain that were based on North American or European lineages of PRRSV, and would not have been detected by the RT-PCR used for screening. Thus, the two RT-PCR-positive detections from one sow and one pen of growers indicate asymptomatic infections with circulating wild-type virus.
Although the infrastructure and laboratory capacity for swine-disease surveillance in Vietnam is limited, government authorities regularly engage in vaccination campaigns for high-priority diseases, and postvaccination monitoring activities afford an opportunity to conduct cross-sectional surveys of viral prevalence. In general, field-based investigations face challenges in obtaining farmer consent for blood collection from pigs, particularly from piglets. Because oral-fluids collections are perceived as posing little or no risk to livestock health, large numbers of diagnostic samples can be easily collected at low cost by staff with limited animal-handling experience. It might be particularly productive to implement oral-fluids collections for case clusters of swine with clinical respiratory disease. We conclude that oral-fluids collection shows promise for future field research on respiratory porcine viruses in Vietnam. However, widespread implementation will require standardization of field sampling techniques and careful adoption of optimized and validated diagnostic assays.
Implications
• PRRSV, IAV, and PCV2 are endemic in swine farms of the Mekong Delta, with moderate levels of PRRSV and IAV transmission and nearly ubiquitous PCV2 circulation.
• Oral fluids provide comparable sensitivity to serum for molecular detection of PCV2.
• Oral-fluids screening can provide an acceptable surrogate for serum samples to estimate overall exposure to porcine respiratory viruses and may prove particularly useful in the context of developing countries.
Acknowledgements
We thank the staff of the SDAH of Can Tho and students of Can Tho University veterinary science department for their assistance in sample collections. This research was supported by the VIZIONS grant of the Wellcome Trust Major Overseas Programme.
Conflict of interest
None reported.
References
1. Cuong VC, Thu NV. Trends in livestock intensification production in Vietnam. Internal report. 2014. Available on request: National Institute of Animal Sciences, Chem, Thuy Phuong, Tu Liem, Ha Noi, Vietnam (http://vcn.vnn.vn/) or by e-mail at Phongdaotao-vcn@gmail.com.
2. Tisdell C. An economic study of small pigholders in Vietnam: Some insights gained and the scope for further research. Working paper No. 61. Economic Theory, Applications, and Issues. University of Queensland, Brisbane, Australia. 2010. Available at: http://results.waterandfood.org/handle/10568/1849. Accessed 25 April 2014.
3. Hoa NT, Chieu TT, Do Dung S, Long NT, Hieu TQ, Luc NT, Nhuong PT, Huong VT, Trinh DT, Wertheim HF, Van Kinh N, Campbell JI, Farrar J, Chau NV, Baker S, Bryant JE. Streptococcus suis and porcine reproductive and respiratory syndrome, Vietnam. Emerg Inf Dis. 2013;19:331–333.
4. Hong TTT, Linh NQ, Ogle B, Lindberg JE. Survey on the prevalence of diarrhoea in pre-weaning piglets and on feeding systems as contributing risk factors in smallholdings in Central Vietnam. Trop Anim Health Prod. 2006;38:397–405.
5. Ngo LT, Hiromoto Y, Pham P, Hong T, Nguyen T, Tri V. Isolation of novel triple-reassortant swine H3N2 influenza viruses possessing the hemagglutinin and neuraminidase genes of a seasonal influenza virus in Vietnam in 2010. Influenza Other Respir Viruses. 2011;2:1–5.
6. Tian K, Xiuling Y, Zhao T, Feng Y, Cao1 Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu Di, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou Lili, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PloS One. 2007 2(6):e526. doi:10.1371/journal.pone.0000526.
7. Metwally S, Mohamed F, Faaberg K, Burrage T, Prarat M, Moran K, Bracht A, Mayr G, Berninger M, Koster L, To TL, Nguyen VL, Reising M, Landgraf J, Cox L, Lubroth J, Carrillo C. Pathogenicity and molecular characterization of emerging porcine reproductive and respiratory syndrome virus in Vietnam in 2007. Transbound Emerg Dis. 2010;57:315–329.
*8. Tung N, Giang N, Takagi M, Inui K, Cam N. Retrospective study on the association of porcine circovirus 2 (PCV2) infections with swine high fever syndrome in Vietnam. Proc Asia Pig Vet Soc Cong. Tokyo, Japan. 2009;245.
9. Nguyen L, Nam H. The study and application of the scientific based strategy for control of PRRS in Viet Nam. Report of the National Project (B1–5PHNC). 2011. Available on request: Department of Animal Health, Epidemiology Division, No. 12, 78th Lane, Giai Phong Road, Phuong Mai, Dong Da, Ha Noi, Vietnam, or by e-mail at nvlong@dah.gov.vn.
10. Creanga A, Nguyen DT, Gerloff N, Hoa TD, Balish A, Nguyen HD, Jang Y, Dam VT, Thor A, Jones J, Simpson N, Shu B, Emery S, Berman L, Bryant JE, Lindstrom S, Klimov A, Donis R, Davis CT, Nguyen T. Emergence of multiple clade 2.3.2.1 influenza A (H5N1) virus subgroups in Vietnam and detection of novel reassortants. Virology. 2013;444:12–20.
11. General Statistics Office of Vietnam. Results of 2011 Rural, Agricultural and Fisheries census. Available at: http://www.gso.gov.vn/default_en.aspx?tabid=477&idmid=4&ItemID=13399. Accessed 24 April 2014.
12. Bhatt S, Lam TT, Lycett SJ, Leigh Brown AJ, Bowden TA, Holmes EC, Guan Y, Wood LN, Brown IH, Kellam P, Pybus OG. The evolutionary dynamics of influenza A virus adaptation to mammalian hosts. Phil Trans Roy Soc B. 2013. doi:10.1098/rstb.2012.0382.
13. Takemae N, Nguyen T, Ngo LT, Hiromoto Y, Uchida Y, Pham VP, Kageyama T, Kasuo S, Shimada S, Yamashita Y, Goto K, Kubo H, Le VT, Vo HV, Do HT, Nguyen HD, Hayashi T, Matsuu A, Saito T. Antigenic variation of H1N1, H1N2 and H3N2 swine influenza viruses in Japan and Vietnam. Arch Virol. 2013;158:859–876.
14. Diep NT, Duong MT, Hoa DT, Huong NT, Tho ND. Serological studies on the circulation pattern of influenza A virus in pig breeding farms in Vietnam. Report, National Center for Veterinary Diagnostics, Department of Animal Health, Hanoi, Vietnam. 2011. Available on request: No. 11, 78th Lane, Giai Phong Road, Phuong Mai, Dong Da, Ha Noi, Vietnam, or by e-mail at ntdiep@dah.gov.vn.
15. Trevennec K, Leger L, Lyazrhi F, Chevalier V, Roger F. Transmission of pandemic influenza H1N1 (2009) in Vietnamese swine in 2009–2010. Influenza Other Respir Viruses. 2012;6:348–357.
16. Trevennec K, Grosbois V, Roger F, Ho TH, Chevalier V. Evidence for freedom from swine influenza in a remote area of Northern Vietnam. Acta Tropica. 2011;10–13. doi:10.1016/j.actatropica.2011.11.012.
17. Ramirez A, Wang C, Prickett JR, Pogranichniy R, Yoon KJ, Main R, Johnson JK, Rademacher C, Hoogland M, Hoffmann P, Kurtz A, Kurtz E, Zimmerman J. Efficient surveillance of pig populations using oral fluids. Prev Vet Med. 2012;104:292–300.
18. Olsen C, Wang C, Christopher-Hennings J, Doolittle K, Harmon KM, Abate S, Kittawornrat A, Lizano S, Main R, Nelson EA, Otterson T, Panyasing Y, Rademacher C, Rauh R, Shah R, Zimmerman J. Probability of detecting porcine reproductive and respiratory syndrome virus infection using pen-based swine oral fluid specimens as a function of within-pen prevalence. J Vet Diagn Invest. 2013;25:328–335.
19. Kottawornrat A, Wang C, Anderson G, Ballagi A, Broes A, Carman S, Doolittle K, Galeota J, Johnson J, Lizano S, Nelson E, Patnayak D, Pogranichniy R, Rice A, Scherba G, Zimmerman J. Ring test evaluation of the repeatability and reproducibility of a porcine reproductive and respiratory syndrome virus oral fluid enzyme-linked immunosorbent assay. J Vet Diagn Invest. 2012;24:1057–1063.
20. Scheltinga S, Templeton KE, Beersma M, Claas E. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J Clin Virol. 2005;33:306–311.
21. Xiao X, Wu H, Yu Y, Cheng B, Yang X, Chen G, Liu D, Li X. Rapid detection of a highly virulent Chinese-type isolate of porcine reproductive and respiratory syndrome virus by real-time reverse transcriptase PCR. J Virol Methods. 2008;149:49–55.
22. WHO Global Influenza Surveillance Network. Chapter 2.I. Molecular identification of influenza isolates. In: Manual for the laboratory diagnosis and virological surveillance of influenza. 2011:83-96. Available at: http://whqlibdoc.who.int/publications/2011/978924158090_eng.pdf.
23. Gall A, Hoffmann B, Harder T, Grund C, Beer M. Universal primer set for amplification and sequencing of HA0 cleavage sites of all influenza A viruses. J Clin Microbiol. 2008;46:2561–2567.
24. Gall A, Hoffmann B, Harder T, Grund C, Ehricht R, Beer M. Rapid and highly sensitive neuraminidase subtyping of avian influenza viruses by use of a diagnostic DNA microarray. J Clin Microbiol. 2009;47:2985–2988.
25. Chittick W, Stensland W, Prickett JR, Strait EL, Harmon K, Yoon KJ, Wang C, Zimmerman JJ. Comparison of RNA extraction and real-time reverse transcription polymerase chain reaction methods for the detection of porcine reproductive and respiratory syndrome virus in porcine oral fluid specimens. J Vet Diagn Invest. 2011;23:248–253.
26. Opriessnig T, Yu S, Gallup JM, Evans RB, Fenaux M, Pallares F, Thacker EL, Brockus CW, Ackermann MR, Thomas X, Meng J, Halbur PG. Effect of vaccination with selective bacterins on conventional pigs infected with type 2 porcine circovirus. Vet Pathol. 2003;40:521–529.
*27. Lam TTH, Duong CM. Detection of porcine circovirus from lesions of postweaning-pig with wasting disease at some farms in Ho Chi Minh City and some adjacent provinces. Proc Workshop Biotech Agric. Nong Lam University, Ho Chi Minh City, Vietnam. 2006.
28. Kittawornrat A, Prickett J, Chittick W, Wang C, Engle M, Johnson J, Patnayak D, Schwartz T, Whitney D, Olsen C, Schwartz K, Zimmerman J. Porcine reproductive and respiratory syndrome virus (PRRSV) in serum and oral fluid samples from individual boars: will oral fluid replace serum for PRRSV surveillance? Virus Res. 2010;154:170–176.
29. Dohoo I, Martin W, Stryhn H. Veterinary Epidemiology Research. Screening and diagnostic tests. Charlottetown, Prince Edward Island: AVC Publishing Inc; 2003;92–126.
30. Olsen C, Karriker L, Wang C, Binjawadagi B, Renukaradhya G, Kittawornrat A, Lizano S, Coetzee J, Main R, Meiszberg A, Panyasing Y, Zimmerman J. Effect of collection material and sample processing on pig oral fluid testing results. Vet J. 2013;198:158–163.
31. Panyasing Y, Goodell C, Wang C, Kittawornrat A, Prickett JR, Schwartz KJ, Ballagi A, Lizano S, Zimmerman JJ. Detection of influenza A virus nucleoprotein antibodies in oral fluid specimens from pigs infected under experimental conditions using a blocking ELISA. Transbound Emerg Dis. 2012. doi:10.1111/tbed.12019.
32. Panyasing Y, Goodell CK, Giménez-Lirola L, Kittawornrat A, Wang C, Schwartz KJ, Zimmerman JJ. Kinetics of influenza A virus nucleoprotein antibody (IgM, IgA, and IgG) in serum and oral fluid specimens from pigs infected under experimental conditions. Vaccine. 2013;31:6210–6215.
33. Kittawornrat A, Prickett J, Wang C, Olsen C, Irwin C, Panyasing Y, Ballagi A, Rice A, Main R, Johnson J, Rademacher C, Hoogland M, Rowland R, Zimmerman J. Detection of porcine reproductive and respiratory syndrome virus (PRRSV) antibodies in oral fluid specimens using a commercial PRRSV serum antibody enzyme-linked immunosorbent assay. J Vet Diagn Invest. 2012;24:262–269.
* Non-refereed references.
