| Brief communication | Peer reviewed |
Cite as: Cappuccio JA, Dibarbora M, Bessone FA, et al. Evaluation of pig pneumonia at slaughter using polymerase chain reaction and histopathology in Argentina. J Swine Health Prod. 2018;26(6):304-308.
Also available as a PDF.
SummaryHistopathology and polymerase chain reaction were conducted on 81 lungs collected at slaughter from 13 swine farms free of porcine reproductive and respiratory syndrome virus and pseudorabies virus infection. Pasteurella multocida and Mycoplasma hyopneumoniae were the most common pathogens detected. Suppurative and catarrhal bronchopneumonia was present in 59 (72.8%) cases. | ResumenSe realizó la reacción en cadena de polimerasa e histopatología en 81 pulmones recolectados en el matadero de 13 granjas porcinas libres del síndrome reproductivo y respiratorio porcino, y de la infección por el virus de la pseudorabia. La Pasteurella multocida y el Mycoplasma hyopneumoniae fueron los patógenos más comúnmente detectados. La bronconeumonía supurativa y catarral estuvieron presentes en 59 (72.8%) casos. | ResuméL’histopathologie et la réaction d’amplification en chaine par la polymérase ont été réalisées sur 81 poumons récoltés à l’abattoir provenant de 13 fermes porcines exemptes du virus du syndrome reproducteur et respiratoire porcin et d’infection par le virus de la pseudorage. Pasteurella multocida et Mycoplasma hyopneumoniae étaient les agents pathogènes les plus communément détectés. Une bronchopneumonie suppurative et catarrhale était présente dans 59 (72.8%) cas. |
Keywords: swine, bronchopneumonia, slaughter, Argentina, porcine respiratory disease complex
Search the AASV web site
for pages with similar keywords.
Received: January 9, 2018
Accepted: June 9, 2018
In pig production systems, slaughter checks are routinely used to estimate prevalence and severity of respiratory disease as well as for detecting subclinical disease. The term porcine respiratory disease complex (PRDC) is used to describe polymicrobial respiratory infections that affect growing and finishing pigs and are associated with economic losses. The etiology of PRDC varies among countries, regions, and farms.1-4 The most common pathogens reported worldwide associated with PRDC are Actinobacillus pleuropneumoniae (Ap), influenza A virus (IAV), Mycoplasma hyopneumoniae (Mh), Pasteurella multocida (Pm), porcine circovirus type 2 (PCV2), porcine reproductive and respiratory syndrome virus (PRRSV), and pseudorabies virus (PRV).1-4 In Asia, Europe, and North America the predominance of PRRSV in cases of PRDC is well documented.2,4
Argentina is free of PRRSV, whereas PRV is under an eradication plan with most confinement farms free of infection. Under this scenario, the agents that are most frequently associated with PRDC in Argentina remain unknown. The aim of this study was to investigate the relationship between respiratory pathogens detected by polymerase chain reaction (PCR) and histopathological lung lesions in lungs with pneumonia lesions obtained from PRRSV- and PRV-free pigs at slaughter.
Materials and methods
All samples were collected from three slaughterhouses in Argentina which operate in accordance with slaughtering procedures approved in the country. A non-probability sampling scheme was applied, only lungs with bronchopneumonia lesions that affected more than 20% of the entire lung area were selected. To avoid cross-contamination in sample processing, sterile instruments were used for each sample collected and samples were individually placed in sterile bags and stored at 4°C. A total of 81 samples were collected from pigs originating from 13 farms (6 to 7 samples from each farm), located in the main swine production areas of the country: Buenos Aires, Entre Rios, and Santa Fe provinces. All 13 farms used the same commercial single-dose, single-injection Mh and PCV2 combined vaccine at the time of weaning (FLEXcombo, Boehringer Ingelheim, St. Joseph, Missouri). No other vaccines against respiratory pathogens were used.
For histopathology analysis, samples were collected and assigned an identification number by a practitioner at the abattoir and then routinely processed according to Laboratorio de Patología Especial Veterinaria procedure manual by a laboratory technician. The prepared blocks were then analyzed in a blinded manner by a single pathologist. Lung lesions were diagnosed into one of the following categories: suppurative bronchopneumonia (SBN) defined by the presence of neutrophils, macrophages, and mucus in bronchioles and alveoli; catarrhal bronchopneumonia (CBN) defined by bronchiole and alveoli filled with mucus exudate, macrophages, and scarce or no neutrophils present; bronchointerstitial pneumonia (BIN) defined by macrophages and lymphocytes infiltrating the alveolar and peribronchiolar septa, bronchiolar necrosis, or hyperplasia of pneumocytes type II; bronchitis and bronchiolitis defined by inflammation or necrosis restricted to airway walls and presence of neutrophils and cellular debris in airway lumen; fibrinous bronchopneumonia (FB) defined by alveoli and interlobular connective tissue filled with serofibrinous exudate, presence of oat cells, and thrombosis of capillaries and lymphatic vessels; or no lesions.
For PCR assays, lung homogenates were processed according to Cappuccio et al.5 Extraction of DNA and RNA was made using Roche High Pure PCR Template Preparation Kit and High Pure RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany). Polymerase chain reaction assays were performed on Veriti Thermal Cycler or StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, California). The presence of IAV, PCV2, Mh, Pm, and Ap were determined as previously described (Table 1).
Table 1: Polymerase chain reaction assays used to measure respiratory pathogens present in lung tissue samples
| Pathogen | Primer sequence | Amplicon size (bp) | Specificity | Type of PCR | Threshold of detection |
|---|---|---|---|---|---|
| IAV* | 5´-GACCRATCCTGTCACCTCTGAC-3´ 5´-AGGGCATTYTGGACAAAKCGTCTA-3´ |
60 | Matrix | RT-PCR | 4.17 × 105 TCID50/reaction |
| PCV2† | 5´- GGGAGGAGTAGTTTACATA-3´ 5´- CGCACTTCTTTCGTTTTC-3´ |
460 | ORF2 | PCR | 4.42 × 105 copies/μL |
| Ap‡ | 5´-GGGGACGTAACTCGGTGATT-3´ 5´-GCTCACCAACGTTTGCTCAT-3´ |
377 | ApxIV | qPCR | 5 CFU/reaction |
| Pm§ | 5´-ATCCGCTATTTACCCAGTGG-3´ 5´-GCTGTAAACGAACTCGCCAC-3´ |
460 | KMT1 | PCR | 4.09 × 103 organisms |
| Mh¶ | 5´-GAGCCTTCAAGCTTCACCAAGA-3´ 5´-TGTGTTAGTGACTTTTGCCACC-3´ 5´-ACTAGATAGGAAATGCTCTAGT-3´ 5´-GTGGACTACCAGGGTATCT-3´ |
649 352 | 16S ribosomal | N-PCR | 80 organisms |
* The presence of IAV was determined as previously described.5
† The presence of PCV2 was determined as previously described.6
‡ The presence of Ap was determined as previously described.7
§ The presence of Pm was determined as previously described.8
¶ The presence of Mh was determined as previously described.9
bp= base pair; PCR = polymerase chain reaction; IAV = influenza A virus; RT-PCR = reverse transcription PCR; TCID50 = 50% tissue culture infective dose; PCV2 = porcine circovirus type 2; ORF2 = open reading frame 2; Ap = Actinobacillus pleuropneumoniae; ApxIV = Actinobacillus pleuropneumoniae toxin IV; qPCR = quantitative PCR; CFU = colony-forming units; Pm = Pasturella multocida; KMT1 = Pasteurella multocida species identification gene; Mh = Mycoplasma hyopneumoniae; N-PCR = nested PCR.
The statistical relationship between frequency of detection of each pathogen and the histopathological category was evaluated by Fisher`s exact test using manual calculation. Statistical significance was set to P < .05.
Results
The most common pathogens detected in 81 samples tested by PCR were Pm in 64 cases (79%) and Mh in 59 cases (72.8%). Our study revealed no PCR positive samples to Ap. Viral pathogens were detected in a lower percentage of samples: PCV2 in 17 cases (21%) and IAV in 6 cases (7.4%) (Figure 1). Coinfections of two or more pathogens were detected in 52 cases (64.2%), with the most common coinfection being Pm and Mh coinfection in 34 of 52 samples (65.4%). In relation to viral pathogens, PCV2 was detected as a coinfection in 15 cases and the 6 IAV positive cases were coinfections.
Figure 1: Frequency of pathogen detection using PCR in 81 lung samples collected at slaughter in Argentina. Single and coinfections are presented. Mh = Mycoplasma hyopneumoniae; PCV2 = porcine circovirus type 2; Pm = Pasturella multocida; IAV = influenza A virus; PCR = polymerase chain reaction.
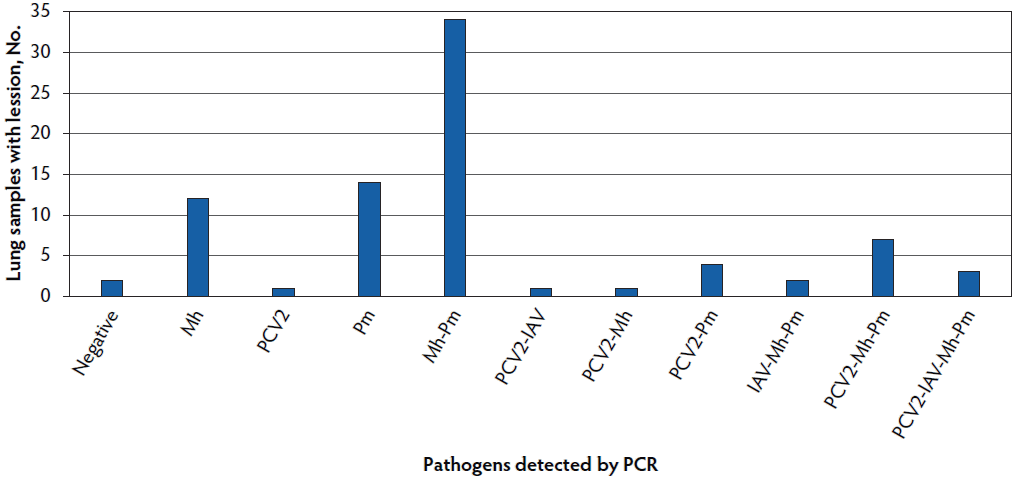
In relation to histopathology analysis, SBN and CBN were present in 59 (72.8%) of the 81 cases (Figure 2) and were most commonly found with either single Mh (7 of 59 cases, 11.9%) or Pm (10 of 59 cases, 16.9%) infections or a combination of both pathogens in 27 of 59 cases (45.8%). Detection of PCV2 occurred in 9 of 34 cases (26.5%) categorized as SBN, of which 3 were also positive for Mh and Pm, 3 were also positive for Mh, Pm, and IAV, 1 was also positive for Pm, 1 was also positive for IAV, and 1 showed no sign of coinfection. Only 3 cases categorized as CBN were positive for PCV2 and were also positive for Mh and Pm (Figure 2). Ten cases were categorized as BIN and tested positive for either Mh, Pm, or both but only 2 cases were PCV2 positive. Only 2 cases were categorized as FB and were negative to Ap and positive to Pm. No statistical association (P > .05) was detected between histopathological classification and pathogen detection by PCR.
Figure 2: Frequency of histopathological categories detected for pathogens present in 81 lung samples collected at slaughter in Argentina. Histopathological categories were: suppurative bronchopneumonia (SBN; 34 samples) defined by the presence of neutrophils, macrophages, and mucus in bronchioles and alveoli; catarrhal bronchopneumonia (CBN; 25 samples) defined by bronchiole and alveoli filled with mucus exudate, macrophages, and scarce or no neutrophils present; bronchointerstitial pneumonia (BIN; 10 samples) defined by macrophages and lymphocytes infiltrating the alveolar and peribronchiolar septa, bronchiolar necrosis, or hyperplasia of pneumocytes type II; bronchitis and bronchiolitis (BB; 8 samples) defined by inflammation or necrosis restricted to airway walls and presence of neutrophils and cellular debris in airway lumen; fibrinous bronchopneumonia (FB; 2 samples) defined by alveoli and interlobular connective tissue filled with serofibrinous exudate, presence of oat cells, and thrombosis of capillaries and lymphatic vessels; and no lesions (NL; 2 samples). Mh = Mycoplasma hyopneumoniae; PCV2 = porcine circovirus type 2; Pm = Pasturella multocida; IAV = influenza A virus; PCR = polymerase chain reaction.
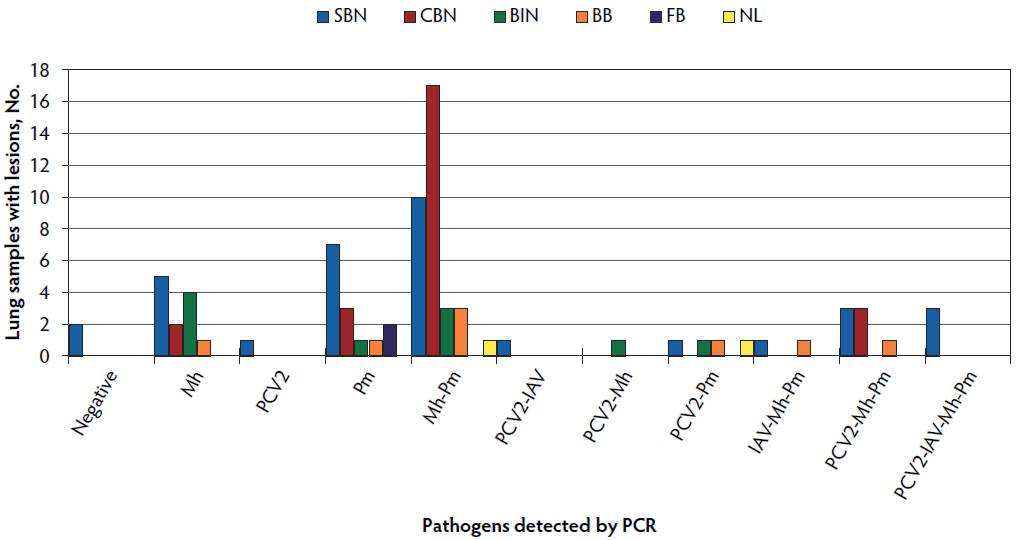
Regardless of histopathological classification, necrotizing bronchiolitis and bronchiolesclerosis were detected in 33 of 81 cases (40.8%), which were most frequently categorized as SBN (23 of 33 cases, 69.7%) and were positive for Mh and Pm (30 of 33 cases, 90.9%). Only 7 cases with necrotizing bronchiolitis and bronchiolesclerosis were positive for PCV2 (5 of 33 cases, 15.2%) or IAV (2 of 33 cases, 6.1%).
Discussion
Few studies have been done to investigate the relationship among pathogen detection and histopathological lesions in PRDC affected pigs.2-4,10 To the best of the authors’ knowledge, this is the first study carried out in Argentina that investigated lung lesions collected from pigs at the time of slaughter. However, it must be taken into consideration, upon interpretation of results, that only a small number of non-randomly selected herds were included in this study and hence, this represents a biased sample of the Argentinian swine population. Regardless of this bias, Pm and Mh were the pathogens most frequently detected. The detection rate of these pathogens is consistent with studies carried out in Asia, Europe, and North America.2-4 The detection of Mh should not be considered a lack of vaccine effectiveness as vaccination reduces clinical signs and lung lesions, but does not prevent the colonization of the organism.10,11 The incidence of viral infections was lower than in previously published studies.2-4 Coinfections of two or more pathogens were detected in a high number of cases (52 of 81; 64%), with the most common being Pm and Mh coinfection. Most of the PCV2 positive cases were coinfections (15 of 17; 88%) further supporting the possible role of PCV2 in coinfections.3,4,10 Influenza A virus was detected in a low number of cases and always in coinfections. The authors hypothesize that the predominance of bacterial detection over viral detections in this study is related to the absence of PRRSV infection and the implementation of vaccination against PCV2 in the farms evaluated but must also reiterate that this was not a randomly generated sample of lung lesions. In the case of IAV, the low detection rate could be affected by a combination of factors including the age of pigs at which the sample was collected, the acute nature of the infection, and the short persistence of the virus in the lungs.2,5
Similar to previous studies,3,5 SBN and CBN were the most common histopathological diagnosis and could be explained by the higher number of samples positive to Pm, Mh, and their coinfection. When CBN or SBN occurs with a bacterial infection without evidence of viral infection, the bronchiolar epithelium is generally normal.12 It is commonly accepted that virus replication leads to inflammation and necrosis of the bronchioles with concomitant obstruction of the lumen that ultimately affects clearance of bacteria and exudates from the alveoli leading to more severe lesions.2,3,12 In this study, necrotizing bronchiolitis or bronchiolesclerosis was detected in 41% of the cases, most frequently associated with SBN and Pm and Mh detection. Bronchointerstitial pneumonia occurs particularly in viral infections and is the more frequent lung lesion associated with PCV2 infections.12 In this study, however, all BIN cases tested positive for either Mh, Pm, or both. Previous studies describe interstitial and peribronchial infiltration lesions to be associated with Mh and Pm infections.12 The 2 cases of FB were positive for Pm and negative for Ap. The relationship between Pm and FB has been previously reported to be associated with toxin production.3 It is highlighted that only one pathologist scored the lung lesions and pathological analysis was performed in a different laboratory than the PCR assays.
The lack of statistical association between histopathological diagnosis and PCR detection of each pathogen could be related to the small number of samples evaluated. Further studies using a more comprehensive epidemiological approach are needed to verify this lack of relationship.
This brief communication reports on the presence of the polymicrobial nature of PRDC in slaughter-aged pigs presumed free of PRRSV and PRV infections and that were vaccinated for Mh and PCV2 at weaning. The presence of bacterial and viral coinfections with SBN lesions in this study supports the continued need to control respiratory infection on swine farms in Argentina. Slaughterhouse inspection of carcasses is used to protect public health by ensuring food safety.13 However, these data collected also have value for other purposes such as passive surveillance activities. For example, the data has been used to estimate prevalence of a particular disease or pathological condition, determine risk factors associated to pleurisy or pneumonia, and to determine the economic effect of pneumonia.14-16 More recently, a well-established slaughterhouse national surveillance system has been considered valuable as an early warning system for an emerging disease or as an initial database to design specific studies (eg, pleurisy or tail lesions).14-17 In this context, slaughter surveys can be applied to generate significant information about the etiology, severity, and interactions of PRDC.
Implications
- Under the conditions of this study, Pm and Mh were the most frequently detected pathogens from grossly affected lungs collected from pigs at slaughter in Argentina.
- This study supports the necessity for the development of a national based slaughterhouse monitoring or surveillance system to continue to document and understand lesions and pathogens present in the Argentinian swine population.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases, Center for Research on Influenza Pathogenesis through the University of Georgia, and Instituto Nacional de Tecnología Agropecuaria on NIAID contract No. HHSN272201400008C. This work was also funded by the Instituto Nacional de Tecnología Agropecuaria Programa Nacional de Sanidad Animal Proyecto Epecífico 1115057; Secretaría de Ciencia y Técnica, Universidad Nacional de La Plata, subsidio V218 and Proyecto de Investigación Científica y Tecnológica 2015-1232, Agencia Nacional de Promoción Científica.
Conflict of interest
None reported.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
1. Opriessnig T, Giménez-Lirola LG, Halbur PG. Polymicrobial respiratory disease in pigs. Anim Health Res Rev. 2011;12:133-148.
2. Choi YK, Goyal SM, Joo HS. Retrospective analysis of etiologic agents associated with respiratory diseases in pigs. Can Vet J. 2003;44:735-737.
3. Hansen MS, Pors SE, Jensen HE, Bille-Hansen V, Bisgaard M, Flachs EM, Nielsen OL. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J Comp Pathol. 2010;143:120-131.
4. Kim J, Chung HK, Chae C. Association of porcine circovirus 2 with porcine respiratory disease complex. Vet J. 2003;166:251-256.
5. Cappuccio J, Dibarbora M, Lozada I, Quiroga A, Olivera V, Dángelo M, Pérez E, Barrales H, Perfumo C, Pereda A, Pérez DR. Two years of surveillance of influenza a virus infection in a swine herd. Results of virological, serological and pathological studies. Comp Immunol Microbiol Infect Dis. 2017;50:110-115.
6. Pereda A, Piñeyro P, Bratanich A, Quiroga MA, Bucafusco D, Craig MI, Cappuccio J, Machuca M, Rimondi A, Dibárbora M, Sanguinetti HR, Perfumo CJ. Genetic characterization of porcine circovirus type 2 from pigs with porcine circovirus associated diseases in Argentina. ISRN Vet Sci. 2011;2011:560905. doi:10.5402/2011/560905
7. Tobias TJ, Bouma A, Klinkenberg D, Daemen AJ, Stegeman JA, Wagenaar JA, Duim B. Detection of Actinobacillus pleuropneumoniae in pigs by real-time quantitative PCR for the apxIVA gene. Vet J. 2012;193:557-560.
8. Townsend KM, Boyce JD, Chung JY, Frost AJ, Adler B. Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J Clin Microbiol. 2001;39:924-929.
9. Calsamiglia M, Pijoan C, Trigo A. Application of a nested polymerase chain reaction assay to detect Mycoplasma hyopneumoniae from nasal swabs. J Vet Diagn Invest. 1999;11:246-251.
10. Fablet C, Marois-Créhan C, Simon G, Grasland B, Jestin A, Kobisch M, Madec F, Rose N. Infectious agents associated with respiratory diseases in 125 farrow-to-finish pig herds: A cross sectional study. Vet Microbiol. 2012;157:152-163.
11. Maes D, Sibila M, Kuhnert P, Segalés J, Haesebrouck F, Pieters M. Update on Mycoplasma hyopneumoniae infections in pigs: Knowledge gaps for improved disease control. Transbound Emerg Dis. 2017;65(Suppl 1):110-124. doi:10.1111/tbed.12677
12. Caswell JL, Williams KJ. Respiratory System. In: Maxie MG, ed. Jubb, Kennedy, and Palmer´s Pathology of Domestic Animals. Vol 2. 6th ed. St Louis, Mo: Elsevier Inc; 2016:465-593.
13. Nielsen SS, Nielsen GB, Denwood MJ, Haugegaard J, Houe H. Comparison of recording of pericarditis and lung disorders at routine meat inspection with findings at systematic health monitoring in Danish finisher pigs. Acta Vet Scand. 2015; 57:18. doi:10.1186/s13028-015-0109-z
14. Brewster VR, Maiti HC, Tucker AW, Nevel A. Associations between EP-like lesions and pleuritis and post trimming carcass weights of finishing pigs in England. Livest Sci. 2017;201:1-4.
15. Jäger HC, McKinley TJ, Wood JLN, Pearce GP, Williamson S, Strugnell B, Done S, Habernoll H, Palzer A, Tucker AW. Factors associated with pleurisy in pigs: A case-control analysis of slaughter pig data for England and Wales. Plos One 2012;7: e29655. doi:10.1371/journal.pone.0029655
16. van Staaveren N, Vale AP, Manzanilla EG, Teixeira DL, Leonard FC, Hanlon A, Boyle LA. Relationship between tail lesions and lung health in slaughter pigs. Prev Vet Med. 2016;127:21-26.
17. Holt HR, Alarcon P, Velasova M, Pfeiffer DU, Wieland B. BPEX Pig Health Scheme: a useful monitoring system for respiratory disease control in pig farms? BMC Vet Res. 2011;7:82. doi:10.1186/1746-6148-7-82
