| Brief communication | Peer reviewed |
Cite as: Erickson AK, Fuhrman M, Mikel WB, et al. Microbiological evaluation of pork offal products collected from processing facilities in a major United States pork-producing region. J Swine Health Prod. 2019;27(1):34-38.
Also available as a PDF.
SummaryAnalysis of 370 offal samples from 15 US pork-processing facilities detected Yersinia enterocolitica-positive (2.4%) and Salmonella-positive (21.8%) samples and mesophilic aerobic plate counts > 107 colony-forming units/g (3.2%). A risk assessment showed intestine (20%), brain (21%), liver and heart (73%), and kidney (87%) sampling batches were acceptable for human consumption. | ResumenEl análisis de 370 muestras de menudencias de 15 centros procesadores de cerdo de EUA detectó muestras positivas al Yersinia enterocolitica (2.4%) y positivas a la Salmonella (21.8%), y conteo de placa aeróbica de mesófilos > 107 unidades/g formadoras de colonias (3.2%). Una evaluación de riesgo mostró que los lotes de muestreo de intestino (20%), cerebro (21%), hígado y corazón (73%), y riñón (87%) eran aceptables para consumo humano. | ResuméL’analyse de 370 échantillons d’abats provenant de 15 établissements de transformation américain a permis de détecter des échantillons positifs pour Yersinia enterocolitica (2.4%) et Salmonella (21.8%) ainsi que des dénombrements de bactéries mésophiles aérobiques > 107 unités formatrices de colonies/g (3.2%). Une évaluation du risque a démontré que les lots échantillonnés d’intestins (20%), de cerveau (21%), de foie et de cœur (73%), ainsi que de reins (87%) étaient acceptables pour consommation humaine. |
Keywords: swine, offal, Salmonella, Yersinia, Toxoplasma
Search the AASV web site
for pages with similar keywords.
Received: March 20, 2018
Accepted: August 21, 2018
Edible offal products from slaughtered hogs represent about 14% of the animal’s live weight.1 These edible offal products include variety meats, which are the edible organs and glands including brain, heart, kidney, liver, thymus gland, and tongue. In the United States, it is estimated that five million metric tons of pork variety meats and other byproducts are generated each year with a large amount of this material being rendered to generate low value products like blood meal, fat, grease, meat and bone meal, and pet food.2 An alternative use of US pork offal would be to market and sell the edible offal products to consumers in countries that prefer strong tasting pork products, like variety meats.3 The desirability of pork offal in foreign markets makes them higher value products in those markets than in the United States, which would likely increase the value of live hogs for US producers.
The purpose of the current study was to determine if pork offal products (brain, heart, intestine, kidney, and liver) as currently produced in US pork-processing facilities are acceptable as food products for human consumption by worldwide populations. To evaluate the microbiological status of pork offal products, sampling batches of five types of pork offal were tested for general contamination and specific human pathogens including Salmonella spp., Yersinia enterocolitica, and Toxoplasma gondii, which have been identified as three of the most common foodborne hazards in pork.4 Salmonella spp. and Y enterocolitica are normal components of the intestinal microflora of healthy pigs that can easily contaminate other pork products within the processing facility environment.5,6 Both Salmonella spp. and Y enterocolitica cause intestinal infections in humans leading to diarrhea.7 Severe Salmonella infections, which occur more commonly in young and elderly persons, can lead to bloody diarrhea, vomiting, and rarely death, while severe Y enterocolitica infections can cause extraintestinal sequelae, such as reactive arthritis, that can persist for years.7 Toxoplasma gondii is a protozoan parasite that can infect a variety of porcine organs including brain, heart, and lungs.8 Toxoplasma gondii causes mild influenza-like symptoms in most infected humans, but it can cause life-threatening infections in fetuses and immunocompromised individuals.9
Materials and methods
The sampling protocol for this study was based on a risk assessment model designed to determine if the offal products coming from an individual processing facility on a particular sampling day are acceptable for human consumption.10 In this model, the two criteria used to design the sampling protocol were: 1) level of concern relative to human health hazards of each potential pathogen (eg, indicator, moderate, serious, or severe) and 2) condition of use of the food product (eg, if the food has a preparation step, such as heating, that would reduce microorganism populations).10,11 The level of risk to humans for the three pathogens evaluated in this study (Salmonella spp., Y enterocolitica, T gondii) is considered serious based on their ability to cause incapacitating, but not usually life-threatening, disease. To be considered acceptable for human consumption when testing for a serious human pathogen with decreased risk due to inactivation by heating, a sampling batch needs to consist of five samples, all of which need to test negative for the presence of the pathogen.10 The mesophilic aerobic plate count (APC) is an indicator test of food acceptability12, with counts less than 1 × 107 colony-forming units/g (CFU/g) considered a negative result. An acceptable APC sampling batch needs to consist of five samples with at least two of the five samples testing negative. Based on this risk assessment model, our sampling protocol included one sampling batch of five types of offal from 15 large pork-processing facilities in ten states (Illinois, Indiana, Iowa, Kentucky, Minnesota, Missouri, Nebraska, North Carolina, South Dakota, and Tennessee) distributed throughout the major pork-producing region of the midwestern and southeastern United States. Selection of slaughterhouses was by convenience, based on proximity to members of the research team. All samples were collected from federally inspected facilities which operate in accordance with the US federal humane slaughter regulations.
Samples of heart, kidney, and liver were obtained from the carcass prior to evisceration or from offal trays depending upon facility operational protocols. An approximately 25-cm segment of ileum was harvested from the intestine just proximal to the ileocecal valve. Brains were harvested by cutting the skull down the median plane using a band saw and then the brain was removed using sterile forceps and placed into a sterile bag. Five samples (> 400 g each) of each type of offal were collected by placing each sample in a sterile Whirl-Pak bag (Nasco, Fort Atkinson, Wisconsin) minimizing cross contamination as best as possible. Offal samples were obtained every 5 to 10 minutes to ensure that animals from multiple farms were represented in each sampling batch. Immediately upon collection, samples were placed on ice and stored at 4°C prior to shipment for laboratory analysis. Tests for Y enterocolitica, Salmonella, and APC were initiated within 96 hours of sample collection. Prior to analysis of the intestine samples, the intestinal contents were removed from the lumen by gently squeezing the contents out of the end of the ileum segment. Approximately 100 g of each offal sample were stored at -20°C for T gondii detection.
Mesophilic aerobic plate counts were performed by homogenizing 25 g of the minced offal sample in 225 mL of buffered peptone water (BPW) using a Seward 3500 stomacher (Islandia, New York) for 2 minutes at 265 rpm. The resulting tissue homogenate was diluted into BPW using 100-fold serial dilutions. One milliliter of each dilution was pipetted onto a 3M Aerobic Count Petrifilm plate (Maplewood, Minnesota) and allowed to incubate at 37°C for 48 hours. Colonies of aerobic bacteria were counted and the APC was calculated as CFU/g of tissue.
Salmonella spp. detection was performed using a method based on the US Department of Agriculture’s Microbiology Laboratory Guidebook.13 Minced offal pieces (25 g) were homogenized in 225 mL of BPW using a stomacher for 2 minutes at 265 rpm and then incubated overnight at 37°C. A commercially available real-time polymerase chain reaction (PCR) method that uses a Hygiena BAX analyzer (Hygiena, Camarillo, California) was used to screen for the presence of Salmonella DNA. Samples that tested positive for Salmonella using PCR were cultured to a pair of selective secondary liquid enrichment media (Hajna Tetrathionate and Rappaport-Vassiliadis Broth; BD, Franklin Lakes, New Jersey) and incubated overnight at 37°C. Ten microliters of each of these broth cultures were spread onto a pair of selective agar plates (XLT and Brilliant Green; BD, Franklin Lakes, New Jersey) and incubated overnight at 37°C. Plates were visually examined to identify potential Salmonella spp. colonies.13 The identity of each suspected Salmonella spp. colony was verified by biotyping using a Bruker Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometer (MALDI-TOF MS; Bruker Daltonics, Billerica, Massachusetts).
For detection of Y enterocolitica, minced offal samples (25 g) were homogenized in 225 mL of BPW using a stomacher for 2 minutes at 265 rpm. One hundred microliters of the resulting homogenate were spread onto MacConkey and Cefsulodin-Irgasan-Novobiocin (CIN) agar plates (BD, Franklin Lakes, New Jersey) and allowed to incubate at 37°C for 48 hours.14 The identity of each suspected Y enterocolitica colony was verified by biotyping using a Bruker MALDI-TOF MS.15
While it is known that T gondii oocysts are partially inactivated by freezing, the DNA-based PCR assay used in this study is capable of detecting the presence of T gondii DNA in frozen tissue.15 Ten grams of minced, thawed offal were placed into a stomacher bag and 25 mL of cell lysis buffer containing 100 mM Tris hydrochloride (pH = 8.0), 5 mM EDTA, 0.2% sodium dodecyl sulphate, 200 mM sodium chloride, and 40 mg/L proteinase K (30 mAnson U/mg) was added. The sample was homogenized using a stomacher at 265 rpm for 2 minutes and then incubated in a water bath at 55°C for 16 hours to release any T gondii oocysts present. The sample was homogenized using a stomacher at 265 rpm for 1 additional minute and then centrifuged for 45 minutes at 3500g. Five milliliters of the supernatant were heated at 100°C for 10 minutes to inactivate proteinase K and then stored at -20°C until PCR testing. The T gondii DNA in the samples was amplified and detected using the primers and real-time quantitative PCR method described by Opsteegh et al.15 A positive control sample of frozen T gondii-infected sheep placenta was used to verify that the sample preparation and PCR methods effectively detected T gondii DNA in frozen tissue samples.
Results
Of the 370 offal samples, 9 (2.4%) tested positive for Y enterocolitica, 81 (21.9%) tested positive for Salmonella spp., 11 (3%) had APC > 107 CFU/g, and 0 (0%) tested positive for T gondii (Table 1). The 9 Yersinia-positive samples included 3 of 70 (4.3%) brains, 1 of 75 (1.3%) heart, 1 of 75 (1.3%) intestine, 2 of 75 (2.7%) kidneys, and 2 of 75 (2.7%) livers. The 81 Salmonella spp.-positive samples included 25 of 70 (35.7%) brains, 9 of 75 (12%) hearts, 37 of 75 (49.3%) intestines, 2 of 75 (2.7%) kidneys, and 8 of 75 (10.7%) livers. All eleven offal samples that had APC > 107 CFU/g were intestine.
Table 1: Percentage of offal samples by type with a positive test for various microbiological pathogens*
| Offal type | Samples that tested positive, No. (%) | |||
|---|---|---|---|---|
| Yersinia enterocolitica | Salmonella spp. | APC > 107 CFU/g | Toxoplasma gondii | |
| Intestine (n = 75) | 1 (1.3) | 37 (49.3) | 11 (14.7) | 0 (0) |
| Heart (n = 75) | 1 (1.3) | 9 (12) | 0 (0) | 0 (0) |
| Kidney (n = 75) | 2 (2.7) | 2 (2.7) | 0 (0) | 0 (0) |
| Brain (n = 70)† | 3 (4.3) | 25 (35.7) | 0 (0) | 0 (0) |
| Liver (n = 75) | 2 (2.7) | 8 (10.7) | 0 (0) | 0 (0) |
| Total (N = 370) | 9 (2.4) | 81 (21.9) | 11 (3.0) | 0 (0) |
* Offal samples were collected from 15 large pork-processing facilities in 10 states distributed throughout the major pork-producing region of the midwestern and southeastern United States.
† One processing plant did not allow the collectors to obtain brain samples.
APC = mesophilic aerobic plate counts; CFU = colony-forming units.
Results from APC analysis of brain, heart, kidney, and liver showed that overall contamination of these types of offal was relatively low with 14 of the 15 facilities having APCs that averaged less than 5.0 log10 CFU/g (Figure 1), which is in the normal range for raw meat samples.12 Average APCs from intestine were much higher than the other types of offal with 10 processing facilities having average APC counts for intestine over 5.0 log10 CFU/g and 6 of those 10 processing facilities having average APC counts for intestine between 6.0 and 7.01 log10 CFU/g. Since none of the sampling batches of offal had 3 of 5 samples with APC counts over 7.0 log10 CFU/g, all offal batches were determined to be acceptable for human consumption based on APC results.
Figure 1: Mesophilic aerobic plate counts of offal samples collected from 15 pork-processing facilities in the United States. Offal tissues sampled were brain, heart, kidney, liver, and intestine. APC = mesophilic aerobic plate counts; CFU = colony-forming units.
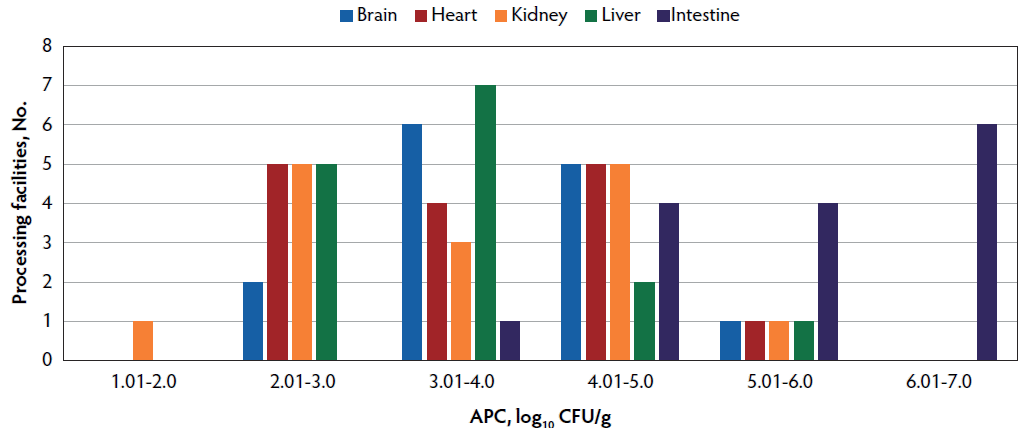
To determine if offal samples produced in a processing facility on a specific day were acceptable for human consumption, the five samples of offal collected from an individual processing facility were considered a sampling batch for risk assessment analysis. In the current study, 68 of 74 (91.9%) sampling batches of all types of offal were acceptable for human consumption based on Y enterocolitica testing and 43 of 74 (58.1%) sampling batches were acceptable based on Salmonella spp. testing (Table 2). All offal sampling batches were acceptable for human consumption based on APC and T gondii testing. All Yersinia-positive samples originated from two processing facilities, so 13 of 15 processing facilities produced five types of offal that were acceptable for human consumption based on Y enterocolitica testing. Salmonella spp. contamination of offal products was much higher with 31 of 74 sampling batches judged unacceptable. These 31 unacceptable sampling batches included 11 brain, 3 heart, 12 intestine, 2 kidney, and 3 liver. For offal coming from a processing facility to be considered acceptable for human consumption, a sampling batch of offal must pass all four microbiological tests. In this study, 41 of 74 (55.4%) sampling batches passed all four tests. Of these 41 acceptable offal sampling batches, only 3 were brain and 3 were intestine. While both brain and intestine are consumed as human foods in various parts of the world, these two types of offal are not as valuable, based on food product desirability and potential export market price,3 as the other three types of offal tested in this study. When we focus on the higher value offal products, which include heart, kidney, and liver, a higher percentage of sampling batches (35 of 45; 77.8%) passed all four microbiological tests and were acceptable for human consumption.
Table 2: Percentage of US pork-processing facilities producing an acceptable sampling batch of each type of offal based on microbiological tests for specific pathogens and APC*
| Offal type | US processing facilities producing a sampling batch of offal acceptable for human consumption, No. (%) | ||||
|---|---|---|---|---|---|
| Yersinia enterocolitica | Salmonella spp. | Toxoplasma gondii | APC | All four tests | |
| Intestine (n = 15) | 14 (93.3) | 3 (20) | 15 (100) | 15 (100) | 3 (20) |
| Heart (n = 15) | 14 (93.3) | 12 (80) | 15 (100) | 15 (100) | 11 (73.3) |
| Kidney (n = 15) | 14 (93.3) | 13 (86.7) | 15 (100) | 15 (100) | 13 (86.7) |
| Brain (n = 14)† | 12 (85.7) | 3 (21.4) | 14 (100) | 14 (100) | 3 (21.4) |
| Liver (n = 15) | 14 (93.3) | 12 (80) | 15 (100) | 15 (100) | 11 (73.3) |
| Total (N = 74) | 68 (91.9) | 43 (58.1) | 74 (100) | 74 (100) | 41 (55.4) |
* Offal samples were collected from 15 large pork-processing facilities in 10 states distributed throughout the major pork-producing region of the midwestern and southeastern United States.
† One processing plant did not allow the collectors to obtain brain samples.
APC = mesophilic aerobic plate counts.
Discussion
The purpose of the current study was to evaluate the extent of microbiological contamination of edible pork offal as currently processed by large US pork slaughterhouses. This study is not intended to be a comprehensive microbiological survey of all types of pork offal from US pork processors. Of the potential foodborne pathogens tested for in this study, Salmonella spp. contamination represents the biggest impediment to marketing US-produced pork offal products as human foods. A similar study of microbiological status of pork offal products produced by Korean slaughterhouses also identified Salmonella as the main foodborne pathogen in pork offal.16 The type of pork offal that was most commonly contaminated with Salmonella spp. was intestine with 12 of 15 sampling batches of intestine determined to be unacceptable for human consumption. Overall, 49.3% of the intestinal samples tested positive for Salmonella spp., which is similar to the percentage of Salmonella-positive cecal samples detected in market swine (35%) and sows (50%) at US slaughterhouses.17 Although it is possible for intestines to become contaminated during processing, the prevalence of Salmonella spp. in this study’s intestinal samples is likely an indication of the percentage of pigs whose intestines (distal ileum) contained Salmonella spp. at slaughter. Since the percentage of intestines that naturally contain Salmonella spp. is high, US pork-processing facilities that want to market intestine as a human food product, such as chitlins, may benefit from incorporating some type of post-harvest disinfection step, eg, an organic acid wash of the intestinal lumen, to decrease levels of Salmonella spp. in these intestinal products.7
The other offal products evaluated in this study, including brain, heart, kidney, and liver, are typically sterile at the time of animal slaughter, but can easily become contaminated by microbes during slaughtering, processing, packaging, and storage.18 The main source of microbial contamination of these offal products in pork-processing facilities comes from tissues, such as intestine, lymph nodes, and tonsils, which are naturally infected with potential foodborne pathogens including both Salmonella and Y enterocolitica.19,20 For example, Salmonella-infected intestine can easily become a source of contamination of other tissues at the time of evisceration of the animal, especially if the intestinal wall becomes perforated and the intestinal contents leak onto other tissues, offal trays, processing equipment, or gloves and tools of facility workers.11 Offal products are particularly vulnerable to this type of contamination since these products are removed from the animal at the same time as the intestine and are then often transported and processed in the same area of the facility as intestine.16
Other than intestine, the pork offal product that was most highly contaminated with Salmonella spp. was brain with 11 of 14 sampling batches determined to be unacceptable for human consumption and 35.7% of brain samples testing positive for Salmonella. The likely reason for the high percentage of Salmonella-positive brains is that the harvesting method resulted in contamination. While other offal samples in this study were obtained from the carcass prior to evisceration or from offal trays, the brain samples were harvested from skulls by splitting the skull down the median plane using a band saw and then the brain was removed and placed into a sterile bag using sterile forceps. The blade of the band saw must cut through multiple types of tissues in the skull including the tonsils, which are known to harbor Salmonella and is a likely source of contamination.20 To effectively market brains as a human food, an alternative method for harvest would have to be implemented to minimize contamination during processing.
Microbiological contamination of heart, kidney, and liver was relatively low in the current study with 10 of the 15 processing facilities having no positive Salmonella spp. results, and 14 of the 15 facilities having no positive Y enterocolitica results. A logical method for further reducing microbial contamination of these types of offal would be to incorporate Hazard Analysis and Critical Control Point (HACCP) systems for processing, packing, transporting, and storing pork offal. The effectiveness of these HACCP programs in reducing contamination of meat products by potential pathogens is demonstrated by 35% of cecal samples from market swine at slaughter testing positive for Salmonella, but only 1.2% of retail pork chops testing positive for Salmonella.17 A similar reduction in the Salmonella contamination of heart, kidney, and liver would likely occur if HACCP systems for pork offal were implemented at all stages of processing.
Implications
- Heart, kidney, and liver as currently harvested by a majority of US processing facilities tested in this study were acceptable for human consumption based on microbiological evaluation for aerobic bacteria, Salmonella spp., Y enterocolitica, and T gondii.
- Of the three potential foodborne pathogens evaluated in this study, Salmonella spp. was the most common contaminant of pork offal products.
Acknowledgements
This research is based upon work supported by a National Pork Board International Trade Research Grant. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the National Pork Board.
Conflict of interest
None reported.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
*1. Marti DL, Johnson RJ, Mathews KH. Where’s the (not) meat? – Byproducts from beef and pork production. https://www.ers.usda.gov/publications/pub-details/?pubid=37428. US Department of Agriculture Economic Research Service report LDP-M-209-01. Published November 2011. Accessed March 29, 2018.
*2. Masker C. Pork variety meat exports add to the producer’s bottom line. Pork Checkoff Report. 2015:34:46.
3. Oh SH, See MT. Pork preference for consumers in China, Japan and South Korea. Asian-Australas J Anim Sci. 2012;25:143-150.
4. Batz MB, Hoffmann S, Morris JG. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J Food Prot. 2012;75:1278-1291.
5. Scallan E, Hoekstra RM, Mahon BE, Jones TF, Griffin PM. An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol Infect. 2015;143:2795-2804.
6. Laukkanen-Ninios R, Fredriksson-Ahomaa M, Korkeala H. Enteropathogenic Yersinia in the pork production chain: Challenges for control. Compr Rev Food Sci Food Safety. 2014;13:1165-1191.
7. Baer AA, Miller MJ, Dilger AC. Pathogens of interest to the pork industry: A review of the research on interventions to assure food safety. Compr Rev Food Sci Food Safety. 2013;12:183-217.
8. Jurankova J, Opsteegh M, Neumayerova H, Kovarcik K, Frencova A, Balaz V, Volf J, Koudela B. Quantification of Toxoplasma gondii in tissue samples of experimentally infected goats by magnetic capture and real-time PCR. Vet Parasitol. 2013;193:95-99.
9. Wang H, Wang T, Luo Q, Huo X, Wang L, Liu T, Xu X, Wang Y, Lu F, Lun Z, Yu L, Shen J. Prevalence and genotypes of Toxoplasma gondii in pork from retail meat stores in Eastern China. Int J Food Microbiol. 2012;157:393-397.
10. International Commission on Microbiological Specifications for Foods. Selection of cases and attribute plan. In: International Commission on Microbiological Specifications for Foods. Microorganisms in Food 7: Microbiological Testing in Food Safety Management. New York, New York: Kluwer Academic/Plenum Press; 2002:145-172.
11. Berends BR, Van Knapen F, Mossel DA, Burt SA, Snijders JM. Salmonella spp. on pork at cutting plants and at the retail level and the influence of particular risk factors. Int J Food Microbiol. 1998;44:207-217.
12. Ryser ET, Schuman JD. Mesophilic aerobic plate count. In: Salfinger Y, Tortorella ML, eds. Compendium of Methods for the Microbiological Examination of Food. 5th ed. Washington D.C.: American Public Health Association; 2015:95-100.
*13. USDA Food Safety and Inspection Service. Isolation and identification of Salmonella from meat, poultry, pasteurized egg, and catfish products and carcass and environmental sponges. In: Microbiology Laboratory Guidebook. Athens, GA: Food Safety and Inspection Service, US Department of Agriculture; 2014:18.
14. Ceylan E. Yersinia. In: Salfinger Y, Tortorello ML, eds. Compendium of Methods for the Microbiological Examination of Foods. 5th ed. Washington D.C.: American Public Health Association; 2015:549-564.
15. Opsteegh M, Langelaar M, Sprong H, den Hartog L, De Craeye S, Bokken G, Ajzenberg D, Kijlstra A, van der Giessen J. Direct detection and genotyping of Toxoplasma gondii in meat samples using magnetic capture and PCR. Int J Food Microbiol. 2010;139:193-201.
16. Im MC, Seo KW, Bae DH, Lee YJ. Bacterial quality and prevalence of foodborne pathogens in edible offal from slaughterhouses in Korea. J Food Prot. 2016;79:163-168.
*17. US Food and Drug Administration. The National Antimicrobial Resistance Monitoring System (NARMS) Integrated Report, 2015. Rockville, MD. US FDA. 2017.
18. Hemmat MI, Reham AA, Omima AS, El Shafay MS. Quality of beef and edible offal at abattoir level. Benha Vet Med J. 2013;25:254-263.
19. Arguello H, Alvarez-Ordoñez A, Carvajal A, Rubio P, Prieto M. Role of slaughtering in Salmonella spreading and control in pork production. J Food Prot. 2013;76:899-911.
20. Chaves BD, Ruiz H, Garcia LG, Echeverry A, Thompson L, Miller MF, Brashears MM. High prevalence of Salmonella in lymph nodes and tonsils of swine presented for slaughter in Mexico. Food Prot Trends. 2017;37:25-29.
* Non-refereed references.
