Original Research (Peer-Reviewed)
Outbreaks of porcine reproductive failure: Report on a collaborative field investigation
H. Scott Hurd, DVM, PhD; Eric J. Bush, DVM, MS; Willy Losinger, MS; Barbara Corso, DVM, MS; Jeff J. Zimmerman, DVM, PhD; Robert Wills, DVM, PhD; Sabrina Swenson, DVM, PhD; Dave Pyburn, DVM; Paul Yeske, DVM; Tom Burkgren, DVM, MBA
HSH: USDA: ARS National Animal Disease Center, Ames IA; EJB, WL, BC: USDA:APHIS:VS Centers for Epidemiology and Animal Health, 555 South Howes, Fort Collins, CO 80526; email: Eric.J.Bush@usda.gov; SS: USDA:APHIS:VS National Veterinary Services Laboratories, Ames IA; JJZ: Iowa State University; RW: University of Nebraska; DP: National Pork Producers Council; PY, TB: American Association of Swine Practitioners
Hurd HS, Bush EJ, Losinger W, et al. Outbreaks of porcine reproductive failure: Report on a collaborative field investigation. J Swine Health Prod. 2001;9(3):103-108. Also available as a PDF.
Please see errata.
Summary
During the latter half of 1996, a series of abortion outbreaks occurred, primarily in southeast Iowa, with a clinical picture similar to Porcine Reproductive and Respiratory Syndrome (PRRS) in a naïve herd. After initial foreign animal disease investigations proved negative, a collaborative case-control study was designed. Its objectives were to describe these cases of intense reproductive failure; attempt isolation of viral agent(s), particularly postulated variant strains of PRRS virus; and identify potentialmanagement risk factors. Eligible case herds were those experiencing a current outbreak of abortion and submitting samples to one of the seven participating diagnostic laboratories. Control herds were randomly selected by the investigating veterinary medical officer from a list generated by the referring practitioner. For both case and control herds, sow blood samples and fetal tissues were collected for PRRSV serologyand multiple virus isolations, and management data were statistically analyzed to identify potential risk factors.
Seventeen case and 34 control herds were enrolled between June 25, 1997 and November 10, 1998. The PRRS virus was isolated from seven cases and one control operation.Sow sera were PRRS positive on 87% of case premises and 55.7% of control premises. These results suggest that the swine abortion outbreaks beginning in 1996 were more likely due to PRRS virus than to an emerging disease. The US pork industry is vulnerable to emerging diseases, and needs to develop rapid capabilities for recognizing new infections and novel pathogens and strategies for detecting and controlling emerging diseases.
Key words : swine, PRRS, epidemiology,
emerging disease
: swine, PRRS, epidemiology,
emerging disease
Received: March 28, 2000
Accepted: January 3, 2001
In 1989, an emerging disease syndrome characterized primarily by reproductive failure was reported in 11 Indiana herds.1 Subsequent reports (1989 to 1990) of the syndrome in other states led the Livestock Conservation Institute to form the Mystery Swine Disease Committee. In October 1990, the Committee developed a case definition for "mystery swine disease." Subsequently, a questionnaire based on this case definition was sent to the membership of the American Association of Swine Practitioners (AASP), with the objective of evaluating the geographic distribution of the syndrome. Among 1305 AASP members in the US, 677 practitioners responded and identified 1611 case herds in 19 states.2
The etiology of this "mystery swine disease" was determined in 1991 with the isolationof the Lelystad virus at the Central Veterinary Institute in the Netherlands.3, 4 A similar enveloped RNA virus was isolated in the US shortly thereafter.5 European workers apparently introduced the terminology "porcine reproductive and respiratory syndrome," or PRRS, circa 1991.
In the US, a retrospective serologic study based on samples collected in Iowa through the National Animal Health Monitoring System (NAHMS) showed that PRRSV entered the Iowa swine population between 1980 and 1985 and spread rapidly.6 The widespread distribution of PRRSV in the US was confirmed by the first NAHMS study in 1990 where at least one of ten sows tested positive in 147 of 412 herds (36%) in 14 of the 17 participating states.7
The clinical effects of PRRSV in naïve herds are highly variable. Infection does not consistently result in clinical disease, and some herds apparently function with few overt problems. Classically, three phases of PRRSV infection in naive herds have been described.8 The initial phase is characterized by inappetance, lethargy, and pyrexia in breeding stock for 1 to 3 weeks, with sow mortality generally less than 3%. During the second phase, respiratory disease appears in young pigs, and a sharp increaseis noted in the number of mummies, stillborn pigs, and weak pigs born to affected sows. Clinical signs are often present for 8 to 12 weeks, after which performanceparameters may return to pre-infection levels. In some cases, a third phase occurs in which endemically infected herds experience chronic disease, particularly respiratory disease in nursery, grower, or finisher pigs.
During the latter half of 1996, a series of abortion outbreaks occurred primarily in southeast Iowa, which presented a clinical picture similar to PRRS in a naïve herd.9 The outbreaks were characterized by abortions in 10 to 50% of sows, regardless of parity, over a 3 to 6 week period. Affected females were anorexic and demonstrated fevers of 40.0 to 41.1 degrees C (104 to 106 degrees F) a few days prior to aborting. Sow death rates were high (5 to 10% of inventory in a 1- to 5-week period). Signs consistent with acute PRRS were occurring in PRRSV-vaccinated herds.10 Outbreaks were also reported in Colorado, Illinois, Nebraska, North Carolina, Minnesota, Missouri, and South Dakota.11
There was concern that these outbreaks represented an emerging disease that could wreak havoc on domestic pork production and export markets. Foreign trading partners were also expressing concern, which initiated a series of collaborative actions by practitioners, laboratory diagnosticians, allied industry, the National Pork Producers Council (NPPC), the USDA Agricultural Research Service, and the USDA Animal and Plant Health Inspection Service (APHIS). At the request of the NPPC and the AASP, APHIS assembled an early responseteam (ERT) to investigate herds affected with abortion outbreaks in southeast Iowa.
The first objective was to quickly rule out the possibility of a foreign animal disease.12, 13 Additionally, the AASP developed a questionnaire and mailed it to practitioners in the United States and Canada, with the purpose of estimating the number of herds meeting the following case definition: abortions in over 10% of sows, including all parities and all stages of gestation, accompanied by mortality in sows and boars exceeding 5% during a 2- to 4- week period.12,14 From this survey, 138 herds meeting the case definition were identified by veterinary practitioners, includingeight herds in Canada.14
The prevailing thought attributed these abortion outbreaks to a severe manifestation of the PRRS virus. However, the question remained whether they were caused by another emerging disease agent, changes in the PRRS virus, or changes in management practices.12,13 Therefore, in late January 1997, an oversight committee of practitioners, producers, veterinary laboratory diagnosticians, pharmaceutical representatives, and animal health officials met to design a collaborative study. Their objectives were to better describe these cases of intense reproductive failure, to attempt to isolate any associated viral agent, and to identify potentialmanagement risk factors. The purpose of this paper is to present preliminary results of the study.
Materials and methods
Farm selection
The study was intended to be a quick-response, case-control study. Cases were to be enlisted as the outbreak progressed. Originally, we had hoped to collect enough data (50 cases, 100 controls) over a 3- to 6-month time frame to complete the study and make recommendations for control. However, fewer cases were identified than anticipated, and data collection was extendeduntil December 1998.
The eligible population consisted of US pork production operations that had at least 50 breeding sows and used a computerized record-keeping system. Eligible case herds were those experiencing a current outbreak of abortion and submitting samples to a participating veterinary diagnostic laboratory (VDL) between June 1, 1997 and December 31, 1998. The seven VDLs were located in Illinois, Indiana, Iowa, Nebraska, North Carolina, Minnesota, and South Dakota. Potential cases were identified by study collaborators at the VDL. These diagnosticians monitored accessions for herds with abortion outbreaks and quickly administered a screening questionnaire by contacting the submitting practitioner. Responses were entered into a secure web data site, maintained by USDA:APHIS: Centers for Epidemiology and Animal Health (CEAH). Using data from the website and conversations with the referring laboratory staff or practitioner, the senior author determined whether the operation qualified for investigation. Case herds were defined as those with abortion rates greater than three per 100 sows in 7 days if the outbreak had occurred within the last 7 days. For outbreaks longer than 7 days, the minimum abortion rate was eight per 100 sows.
Control herds were randomly selected from a list of ten candidate herds generated by the practitioner who referred the case herd. Cooperating practitioners were visited by their local Federal Veterinary Medical Officer (VMO) who further explained the study and helped with control herd selection. The goal was to obtain three qualifying control herds for each case. Potential control herds were screened with the same questionnaire used for screening case herds. Control herds were not accepted if they did not meet the selection criteria or reported a previous episode of severe PRRS with abortions > 5% per month or preweaning mortality > 25% per month for any month in the previous 15. This definition did not exclude herds that were endemically infected with PRRSV. Case and control herds were given a numeric identification to maintain the confidentiality of the producer. Only the visiting VMO knew both the producer name and numeric identification.
Data collection
Farm visits were jointly conducted by the herd practitioner and VMO for both case and control herds. The VMOs attempted to visit case herds within 7 days of the initialreport to CEAH, to complete a management questionnaire and review summary production reports. The questionnaire included questions on a wide range of management-related topics, including biosecurity practices such as sourcing of replacement animals, showering, perimeter fencing, off-site weaning or finishing, and non-employee access to facilities.Data regarding disease and vaccination history were collected, including brand-specific history of PRRS vaccines used. Housing data included flooring type in gestation, farrowing, nursery, and finishing barns, animal density in shared airspace, and type of watering system in each building. A copy of the questionnaire is available from the CEAH upon request.
Sample collection
Blood samples were collected from 30 randomly selected sows or gilts in case and control herds. In addition, blood samples were collected from recently aborted sows or gilts. Sera from these samples were assayedfor PRRSV and PRRSV antibodies. Convalescent blood samples were collected from the same animals approximately 3 weeks later, and re-assayed for PRRSV antibodies. Tissues (lung, liver, kidney, and spleen) from recently aborted fetuses or newborn piglets were also collected at the first visit and tested for viruses.All samples were submitted to the Diagnostic Virology Laboratory at the National Veterinary Services Laboratories, Ames, Iowa. Foreign animal diseases (classical swine fever, African swine fever, foot-and-mouth disease) had already been ruled out by diagnostic laboratory work and the ERT. Therefore, detailed diagnostics focusedon virus isolation and PRRSV serology.
Serology and virus isolation
Sera were tested for PRRSV antibodies by ELISA using the HerdChek(R) Porcine Reproductiveand Respiratory Syndrome Virus Antibody Test Kit (IDEXX Laboratories, Westbrook, Maine), according to the manufacturer's instructions.
A 20% tissue homogenate was prepared for each piglet tissue submitted. Homogenates, sera, and thoracic fluids were prepared for MARC-145 cell culture inoculation by passing them through a 0.45 µm filter. InoculatedMARC-145 cells were observed for 9 to 11 days and then blind passaged. Cell cultures showing cytopathic effects (CPE) were subpassaged and stained with anti-PRRSV monoclonal antibodies SDOW17 and V017. Cell cultures in which no CPE was observed after the second passage (7 days) were reported as virus isolation negative.
Tissue homogenates were also inoculated on primary fetal swine kidney cells. Cell cultures showing CPE were subpassaged and stained with a polyvalent porcine viral antiserum for detection of encephalomyocarditis virus, hemagglutinating encephalomyelitis virus, porcine adenovirus, porcine parvovirus, reovirus, rotatvirus, pseudorabies virus, transmissible gastroenteritis virus, H1N1 swine influenza virus (SIV), and porcine enteroviruses (groups 1 to 8c). Cell cultures in which CPE were not observed at the end of the incubation period were subpassaged and stained with the polyvalent porcine viral antiserum.
Virus isolates identified in MARC-145 cells were further propagated on MARC-145 cells several times until consistent CPE was observed. The bulk fluids were aliquoted for distribution and tested for extraneous agents by passing in porcine kidney 15 (PK-15), bovine turbinate (BT), swine kidney, and swine testicular cell cultures. The PK-15 cultures were stained with classical swine fever conjugate and the BT cultures were stained with a polyclonal bovine diarrhea antiserum. The bulk fluids were also inoculated on bovine blood agar plates to screen for miscellaneous aerobic bacteria, and tested for mycoplasma accordingto federal guidelines15 for vaccine sterility. Viral isolates were made available to interested researchers and can be obtained by contacting the CEAH.
Data analysis
Completed questionnaires were sent to the CEAH and edited for completeness and consistency. Unclear items were referred to the VMO who had completed the questionnaire. Data were entered into EpiInfo (USD Inc., Stone Mountain, Georgia), a database developed by the Centers for Disease Control for use in field disease outbreaks. Serologic and production data were analyzed by analysis of variance using Statistix (Analytical Software Inc, Tallahassee, Florida).
Management and production data from the questionnaire were
analyzed using the original case and control herd definitions.
The differences in production data between case and control herds
were evaluated with a two-sample t-test, assuming unequal variances.
All management variables were evaluated, with case or control
as the dependent variable. The probability of being a case was
evaluated by calculation of an odds ratio (OR) and chi-square
statistics using EpiInfo. After initial two-by-two screening of
all variables, variables with a
P <.2 were offered to a logistic regression modeling
procedure using a forward stepwise selection in SAS LOGISTIC (SAS
Institute, Cary North Carolina). After multiple iterations of
this stepwise procedure, a near final model was defined. Finally,
the change in the adjusted OR resulting from the removal of any
one remaining variable from the modelwas evaluated. Additionally,
the effect on model precision of removing any one variable was
evaluated with the likelihood ratio test. This method is recommended
over evaluating the P value for the regression coefficient.16
The final model reflects the association of each variable with
the probability of a herd being a case, while adjusting for the
remaining variables in the model.
Results
Seventeen case herds and 34 control herds were enrolled between June 25, 1997 and November 10, 1998. One control herd was excluded from analysis because it appeared to be experiencing an outbreak of acute PRRS when the VMO arrived to collect convalescent samples. The number of control herds identified for each case herd varied from zero to three. Nine of the herds had three associated controls, one had two controls, and five had one control. For two case herds, no control herd was identified. Most case herds (ten herds, 58.8% of cases) and control herds (20 herds, 60.6% of controls) were provided by Iowa's VDL contacts and practitioners. Other states provided fewer case and control herds. Illinois and Minnesota each provided one case and three controls, Nebraska provided three cases and five controls, and North Carolina provided two cases and two controls. Enrolledherd types for case and control herds combined were farrow-to-finish (22 herds), producers of weaned pigs (21 herds), feeder pigs (six herds), or seed stock (two herds). All but seven farrowing sites were managed as AI-AO, either by room (32 herds) or by building (11 herds).
For the 17 case herds, the breeding inventory ranged from 105 to 2495 sows and gilts, with a median of 610 breeding females.For the 33 control herds, the breeding inventory ranged from 65 to 3677 sows and gilts, with a median of 556 breeding females. All case herds except one were single site operations. Five of the control herds had multiple breeding sites while the other 28 were single site operations.
Except for one control herd, all enrolled herds had exposure to PRRSV. A diagnosis of PRRS had been made on 13 case herds between September 1987 and January 1998, and on 20 control herds between July 1990 and March 1998. At some time in the past, PRRS vaccine had been used in 15 case herds (88%), but at the time the samples were collected, only nine case herds were using PRRS vaccine. Vaccine had been used in 24 of the control herds (72%), but at the time samples were collected, only 16 control herds were using PRRS vaccine. There was no significant difference in the distribution between case and control herds for any of the above management and PRRSV exposure variables.
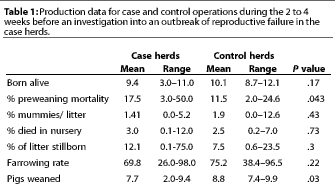 Table 1 shows a summary of the
reported production data for case and control premises in the
2 to 4 weeks preceding the investigation. Case premises had higher
(P=.043) preweaning mortality (17.5%) in the 2 weeks before
investigation, compared to control premises (11.5%). Case premises
weaned fewer (P=.03) pigs (7.7) than control premises (8.8).
No other production parameters appeared significantly different
between case and control herds.
Table 1 shows a summary of the
reported production data for case and control premises in the
2 to 4 weeks preceding the investigation. Case premises had higher
(P=.043) preweaning mortality (17.5%) in the 2 weeks before
investigation, compared to control premises (11.5%). Case premises
weaned fewer (P=.03) pigs (7.7) than control premises (8.8).
No other production parameters appeared significantly different
between case and control herds.
Using samples collected by the VMO, PRRSV was isolated from eight different herds (seven cases, one control). Overall, PRRSV was isolated from 33 different samples: 26 sera, six piglet tissues, and one fetal thoracic fluid. Virus was isolated from five tissue samples and ten serum samples from one of the case farms. For the one control herd from which PRRSV was isolated, the two isolations were from sow sera. No other viruses were detected in any of the samples submitted.
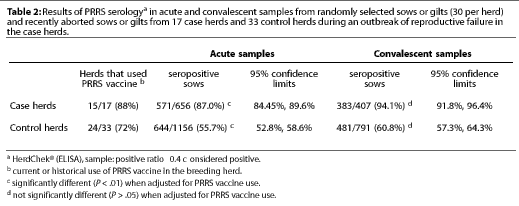
The sow serological results are shown in Table 2. A sample was considered seropositive if the sample-to-positive ratio was 0.4. On the first collection, 87% of serum samples from case herds and 55.7% of samples from control herds were positive. The average percent of sows positive differed (P <.01). between case and control premises, even when adjusted for the historical or current use of PRRS vaccine in the breeding herd. For both case and control premises, the percent of PRRS-positive sows tended to be greater for the convalescent samples than for the acute samples; however, these increases were not statistically significant (P>.05) when adjusted for PRRS vaccine use as a covariant.
Variables which passed the initial two-by-two comparison and were offered to the stepwise multivariate logistic regression procedure included removing feeder pigs to a separate site, full slats or mesh floor in farrowing building, trough water in the farrowing facility, livestock trucks from other herds allowed on site, non-acute PRRS in the previous 12 months, swine influenza in the previous 12 months, use of any PRRS vaccine, sourcing of new females or males from PRRS-positive or status-unknown sources, and isolation of PRRSV during the investigation. A herd was recordedas positive for non-acute PRRS if it had been diagnosed by a veterinarian's opinion or was positive on any of the available laboratory tests, including virus isolation. Of the 32 herds reporting a previous PRRS diagnosis, ten reported an on-farm veterinary diagnosis as the only source. Reporteduse of a computer service bureau was associated with decreased risk on the two-by-two comparison. This variable was also offered to the modeling procedure. Questions regarding general and brand specific PRRS vaccine use were evaluated by two-by-two comparison with no observed effect on the risk of acute PRRS.
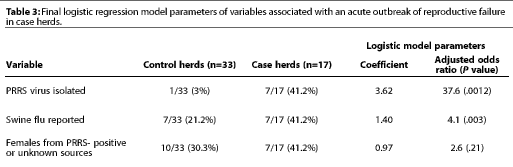
Results of the final logistic regression model are shown in Table 3. Case herds were much more likely than control herds to have the PRRS virus on the premises at the time of investigation (OR=37.6). Additionally, having experienced an outbreak of swine influenza in the previous 12 months correlated with an increased risk (OR = 4.1). Lastly, receiving gilts from outside sources that were "PRRS positive or source-unknown" correlated with an increasedrisk (OR= 2.6).
Discussion
Results of this study suggest that the swine abortion outbreaks beginning in 1996 were not caused by an emerging disease, but rather by the PRRS virus. Only one control operation yielded the PRRS virus, compared to seven of the case herds. However, it must be noted that no attempt was made to diagnose porcine circovirus type II. The case-control study design was a valuable approach, as data collection from case premises alone would not have clearly demonstrated the role of PRRSV. For whatever reason, these case herds did not have a protective level of herd immunity. It is not clear if cases resulted from the introduction of a variant PRRSV, the short duration of immunity from natural exposure, or a vaccine failure. A recent study of monoclonal antibody patterns of the virus from this outbreak suggested that these isolates differ antigenically from previous isolates.17 Isolates from this study have been provided to other researchers, whose work should further clarify viral strain issues.
The observation of an increased risk of acute PRRS with recent SIV experience suggests a possible synergistic role between the two agents. It is possible that SIV was a major contributor to the clinical syndrome. However, no SIV was isolated from these herds. Had SIV of any type been present in the samples collected, our virus isolation methods would have identified it. Synergism between PRRSV and other pathogens has been reported elsewhere. Swine influenza virus, chlamydia, Hemophilus parasuis, Actinobacillus pleuropneumonia , Pasteurella multocida, Pasteurella haemolytica, and Streptococcus suis respiratory infections have been recognized in association with PRRSV infections.17,18 Increases in the incidence and severity of salmonellosis, Escherichia coli, and Clostridia infections, polyserositis, greasy pig disease, atrophic rhinitis, swine dysentery, sarcoptic mange, and S suis meningitis were also noted. Actinobacillus pleuropneumoniae, swine influenza virus, encephalomyocarditis virus, pseudorabies virus, porcine cytomegalovirus, porcine respiratory coronavirus, and porcine paramyxovirus are common secondary infections reportedly associated with herds chronically infected with PRRSV.19 Investigators demonstrated a positive association between seroprevalence of PRRSV and porcine respiratory corona virus, and between PRRSV and porcine influenza virus strain Arnsberg subtype H1N1.19 The risk of bringing females into the herd from PRRSV-positive sources highlights the need for increased biosecurity. This point is particularly important, as new virus strains might be introduced.
In this paper, we reported on the effects of various management factors using the original case definition. However, these management factors could be evaluated with alternative case definitions (dependent variables) created from the production data. The entire dataset is available for analysis by contacting the CEAH.
Because of the low incidence of outbreaks meeting the case definition, enrollment in this study was lower than originally anticipated and was extended to December 1998. The expected study size fell short of the planned 50 case herds and 100 control herds. The low case enrollment may reflect a number of factors, such as a decrease in the incidence of acute porcine reproductive failure and (or) unwillingness of producers or practitioners to participate in the study. Lack of understanding about the various roles of producers, practitioners, and VMOs may have led to a lack of trust regarding confidentiality of data, especially in light of the exceptional level of media attention surrounding the early episodes of abortion outbreaks.
In the early stages of this outbreak, the syndrome was called Sow Abortion and Mortality Syndrome (SAMS), "Atypical PRRS," or "Hot PRRS". Although case producers originally complained of high production losses, this investigation found that clinical losses were not as severe as first reported, and that the clinical picture actually fit the losses previously described in herds newly affected with the PRRS virus (Table 1). For this reason, the term "acute PRRS" seemed appropriate. This terminology avoided the impacts on international trade generated by any inference about excessive severity of the syndrome and any suggestion that the virus was different from previous PRRSV isolates.
Experiences during the outbreak and the difficulties with implementing the collaborative study exposed an Achilles' heel in the vibrant US pork industry. Pork producers increasingly rely on pork production as their sole source of income, and the pork industry is increasingly dependent on the expanding export market for its growth. The mere rumor of an emerging disease can close export markets. Even the name attached to the emerging syndrome has trade impacts. Additionally, if a highly contagious and pathogenic disease does occur, production or animal losses can be significant. The sudden disappearance of export markets in combination with disease losses places pork producers at risk of losing their livelihood.20
Currently, control of emerging diseases seems to fall between the cracks of the pork industry's health infrastructure. Practition-ers typically focus on disease control at the herd and county level. The APHIS Veterinary Services is traditionally involved in disease control for "program diseases" or in responding to foreign animal disease threats. There is no mechanism in place to recognize and respond to an emerging disease in the US pork industry.20 The implications of a poor response to an emerging disease are not widely appreciated, but can be clearly demonstrated by the recent situation in Malaysia with the Nipah virus.21 Additionally, the devastating effects of a highly contagious disease are being demonstrated with foot-and-mouth disease in Europe.
This acute PRRS investigative study represents a first attempt for an innovative, coordinated response to a possible emerging disease occurring in multiple counties and states. Attempts for a coordinated response were awkward, but instructive. Two previously described lessons were immediately evident. First, the pork industry needs to develop the capability to rapidly identify and contain an emerging animal disease, and, second, the planned rapid response for foreign animal diseases is not an appropriate paradigm for responding to emerging diseases.22, 23 The response demonstrated in this collaborative study is new territory for allied pork industry. It represents an important first step. However, numerous deficiencies surfaced regarding collective abilities to respond to an emerging disease.
Implications
- The 1996-97 outbreak of reproductive failure resembled PRRSV infection in naïve herds and was likely caused by the PRRS virus.
- The US pork industry needs to develop capabilities for rapid recognition and evaluation of new disease patterns.
- Pork producers, swine practitioners, and allied industry must formulate plans to effectively and cooperatively respond to future emerging diseases in a timely fashion.
- Pork producers, swine practitioners, and allied industry appear willing to cooperate at some level in addressing concerns about an emerging disease if the confidentiality of the producer can be maintained.
Acknowledgements
Special thanks to the diagnosticians, producers, practitioners, and APHIS veterinary medical officers who participated and contributed to the field data collection.
References -- refereed
3. Terpstra C, Wensvoort G, Pol JMA. Experimental reproduction of porcine epidemic abortion and respiratory syndrome (mystery swine disease) by infection with Lelystad virus: Koch's postulates fulfilled. Vet Q. 1991;13:131-136.
4. Wensvoort G, Terpstra C, Pol JMA, et al. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q. 1991;13:121-130.
10. Halbur PG, Bush EJ. Update on abortion storms and sow mortality. Swine Health Prod. 1997;5:73.
14. Zimmerman JJ, Epperson W, Wills RW, McKean JD. Results of the recent survey of the membership of the AASP for outbreaks of sow abortion and mortality. Swine Health Prod. 1997;5:74.
15. Animals and Animal Products, 9 CFR S28 (1998).
16. Kleinbaum D, Kupper L, Morgenstern H. Epidemiologic Research, Principles and Quantitative Methods. New York: Van Nostrand Reinhold Company, 1982;430-432, 455.
17. Done SH, Paton DJ. Porcine reproductive and respiratory syndrome: clinical disease, pathology and immunosuppression. Vet Rec. 1995;136:32-35.
19. Groschup MH, Brun A, Haas B. Serological studies on the potential synergism of porcine reproductive and respiratory syndrome virus and influenza-, corona- and paramyxoviruses in the induction of respiratory symptoms in swine. Zentralbl Veterinarmed B. 1993;40:681-9.
21. Centers for Disease Control and Prevention. Outbreak of Hendra-Like Virus -- Malaysia and Singapore, 1998-1999. MMWR. 1999;48:265-269.
References -- non-refereed
1. Keffaber KK, Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsletter. 1989;1:1-10.
2. Zimmerman JJ. A survey of the American Association of Swine Practitioners for the presence of mystery swine disease herds. Proc Livest Conserv Inst. Bloomington, Minnesota. 1991;121-128.
5. Collins JE, Benfield DA, Christianson WT, et al. Swine infertility and respiratory syndrome (mystery swine disease). Minn. Swine Conf Vet. St. Paul, Minnesota. 1991;200-205.
6. Hill HT, Owen WJ, Eernisse KA, Zimmerman JJ, Uhlenhopp EK, Frey ML. Prevalence of SIRS in Iowa swine herds. Proc Int Symp SIRS/PRRS/PEARS. 1992;35.
9. AASP PRRS Subcommittee. Recent PRRS Outbreaks. AASP Update. January 1997.
11. Epperson W. An Abortion Storm and Sow Mortality Syndrome. AASP 28th Annual Meeting. Quebec City, Quebec. 1997;479.
12. Hurd HS. The Acute PRRS Investigative study - an update. George A. Young Swine Conf Ann Nebraska SPF Swine Conf. Lincoln, Nebraska. 1997;143.
13. Bush EJ. PRRS Epidemiology Team Report. LCI Ann Meet. Colombus, Ohio. 1997;117.
17. Yang L, Nilubol D, Harris DL, Platt KB. Proc Conf Res Work Anim Dis. 1999;171.
18. Joo HS, Dee SA. Recurrent PRRS problems in nursery pigs. Allen D Leman Swine Conf, St. Paul, Minnesota. 1993,20:85-86.
20. Lautner B. Swine Futures Project- Partnership for Progress. Proc 101st Ann Meet US Anim Health Assoc. Louisville, Kentucky. 1998;520-523.
22. Bush EJ. Surveillance for emerging diseases associated with swine: An update on the Swine Futures project. LCI Ann Meet. Des Moines, Iowa. 1998.
23. Lautner B. Emerging diseases - Are there alternatives to the "vet rumor mill"? Swine Health Prod. 1998;6:245-6.
