Diagnostic notes |
Non refereed |
Serologic basis for assessment of subclinical Salmonella infection in swine: Part 2
Isabel Turney Harris, DVM, PhD
Department of Microbiology, College of Agriculture, Iowa State University, Ames, Iowa. Address correspondence to: Dr Isabel Turney Harris, Department of Microbiology, 207 Science I, Ames, IA 50011; Tel: 515-294-7058; Fax: 515-294-6019; E-mail: iharris@iastate.edu.
Cite as: Harris IT. Serologic basis for assessment of subclinical Salmonella infection in swine: Part 2. J Swine Health Prod. 2003;11(6):300-303.
Also available as a PDF.
Search the AASV web site for pages with similar keywords.
This is Part 2 of a two-part series. In Part 1, topics included control of subclinical salmonellosis in swine, ELISA tests to detect Salmonella serum and meat juice antibodies and the sensitivity and specificity of these tests, national Salmonella surveillance programs, serological tests used in the monitoring programs, and correlation of serological test results with culture results.
Detection of serotypes of importance
For optimum sensitivity, the Salmonella ELISA should incorporate antigens capable of detecting antibodies to the predominant serotypes in the geographical area where animals are to be tested.1 The Danish mix-ELISA (DME) detects O antigens from about 93% of isolates found in pigs in Denmark.2 In the United States, a study in which pooled pen fecal samples from 37 farms were cultured resulted in 286 Salmonella isolates, 92% containing O antigens capable of producing antibody detectable by the DME.3 In the report of serotypes most frequently isolated from swine by the National Veterinary Services Laboratory (Ames, Iowa), 91% were in O antigen groups included in the DME.4 When Salmonella is isolated on a premise, multiple serotypes are usually identified.3
Herd test compared to individual test
At its present level of sensitivity and specificity, the Salmonella ELISA is applied in the field as a herd test, not an individual animal test. The response of the individual animals sampled are evaluated in order to make a decision on the status of the whole herd.5 Therefore, at any sampling time point, pigs in the sampled group may be at different stages in their serologic responses, depending upon when they were exposed to Salmonella, the infective dose, and the degree of immunologic response detectable by the test. The results of the individual test determinations are pooled to make an interpretation on the status of the herd. Evaluation of the herd as the unit is useful because infection levels may vary greatly between groups. Clinical trials on the effectiveness of intervention strategies designed to reduce subclinical infection are necessarily conducted on groups. To be valid, methods of intervention must be tested and applied to groups, not individuals, because of the dynamics of Salmonella infection in the herd.6
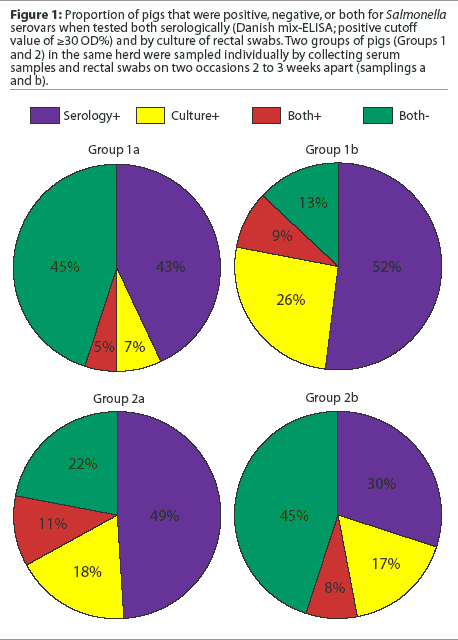
Several studies have reported the variability of serologic responses in different groups of animals and within groups of animals sampled at the same time and at different times. In one study, seroprevalence ranged from 0 to 80% in individual buildings at a single locus in a multi-site system.7 In another study, in which two groups of pigs from three different herds were sampled twice at a 2- to 3-week interval (I. T. Harris, unpublished data), the proportion of animals that were positive by culture, serology, or both varied between groups and within groups of pigs sampled at different times (Figure 1). Between collection of the first and second samples in the first group of pigs, the proportions of positive serologic and cultural results both increased. In the second group of pigs, the proportion of seropositive animals decreased between the first and second samplings, while culture results remained about the same.
Differences in experimental and field use of ELISA tests
Salmonella ELISA tests are useful as individual pig tests in research situations, where pigs are experimentally infected with pure cultures of known serotypes of Salmonella.The "cutoff" selected to distinguish positive and negative results may be different for the test under research conditions compared to the same test used in field studies because of potential variations in the time when the pigs are exposed to Salmonella, the infective dose, and the degree of immunologic response detectable by the test.8 Since the color change read as optical density by the ELISA reader is a continuous variable, test samples are compared with control samples of known antibody concentration, or known reactivity or lack thereof. Control samples are usually produced by experimentally infecting animals with pure cultures of known serotypes of Salmonella and collecting their sera. Likewise, sensitivity and specificity determinations are made by comparing serological results to cultural results of swabs collected from the same experimentally infected animals, if culture is used as the "gold standard." Sensitivity and specificity may be quite different in a field situation compared to research conditions. The animals might have antibodies to other Entero-bacteriaceae that cross react with components of the ELISA; the infectious dose of Salmonella may be low (i.e., <=103 colony forming units); there may be Salmonella serovars of varying antigenicity; there may be varying levels of immunocompetence in the animals exposed; there might be multiple serovars of Salmonella on the farm, or serovars containing antigens not included in the ELISA; or there might be an effect on the antibody response of concurrent infections in the herd or vaccine or antibiotic use in the herd. It has been reported that an avirulent vaccine used to prevent salmonellosis in pigs due to Salmonella serovar Choleraesuis does not induce antibodies detectable by the DME.9
Repeated testing in serologic surveillance
The concept of serologic testing is most useful as an ongoing monitoring surveillance exercise. The sensitivity of the test increases with multiple samplings. A single determination of seroprevalence may not accurately represent the status of a herd. The serologic response to subclinical Salmonella infection may vary between groups of pigs in a finisher or even within the same group sampled at different times.10,11 Variations in infection may be related to the number of pigs infected or to the infectious "load" they are carrying or exposed to in their environment. Certain buildings may have characteristics that allow for contamination to build up, including some types of flooring, waste disposal methods, feed delivery systems, and materials that are difficult to clean and disinfect. Rodent and insect control, heating and ventilation variations, and seasonal effects all may relate to environmental exposure. Repeated determination of seroprevalence in different groups of pigs may provide a more accurate assessment of a herd's subclinical infection status and establish a baseline for the herd. Repeated testing on a periodic (monthly) basis may reveal seasonal and environmental effects, as well as variations in the degree of Salmonella exposure or in the Salmonella serovars present in the groups of animals tested.12,13
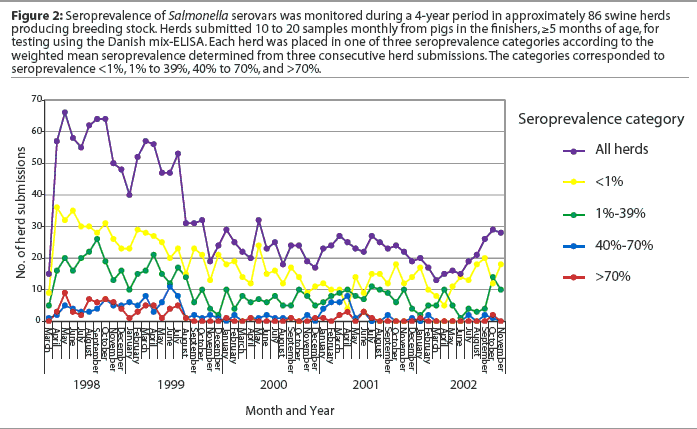
The DME was designed for surveillance and is recommended for monitoring herds and detecting high levels of Salmonella infection.8Our laboratory began a project in 1998 using the DME to monitor subclinical Salmonella infection in a group of herds producing breeding stock. The procedure was based on the Danish system for monitoring Salmonella in breeding stock producers.14 Briefly, 10 to 20 serum samples collected monthly from animals in the finishers, 5 months of age or older, are assayed with the DME. The Salmonella index is calculated from the weighted mean of seroprevalence determinations from three consecutive herd submissions. The calculated index places the herd in one of three seroprevalence categories, with the lowest index named Level 1, as in the Danish control program. Results of the first 4 years of testing, showing the number of herd submissions falling within each range of seroprevalence, are illustrated in Figure 2. Among the 1678 submissions from approximately 86 herds, we have found very few submissions with seroprevalence >40% (I. T. Harris, unpublished data).
Possible intervention strategies of merit
Field studies have identified several procedures that reduce Salmonella seroprevalence. Basically, these encompass all in-all out practices, cleaning and disinfection between batches, and strict control over the introduction of external sources of Salmonella (in pigs or feedstuffs).8 Salmonellae may survive up to 3 months in wet feces and 13 months in desiccated material.15 The use of home-ground barley feed or fermented liquid feeds, the addition of organic acids to the feed, or all three, may reduce Salmonella seroprevalence in groups of pigs.16-20 Vaccination for Salmonella Choleraesuis in the United States, not widely practiced in other countries (for example, in Denmark, where Salmonella Choleraesuis does not occur), may reduce the occurrence of subclinical salmonellosis.9, 21-26 The feeding of home-ground corn-soy rations, which is common in the United States but not in other countries, may account for differences in the prevalence of subclinical infection between US herds and herds in other countries. There is some evidence that subclinical Salmonella infection may affect the growth performance of swine. A study which compared growth in groups of pigs with different Salmonella seroprevalence levels showed that low seroprevalence groups gained more than pigs in high seroprevalence groups.9 High Salmonella seroprevalence may be an overall indicator of cleanliness, effectiveness of rodent control, stocking density, and other management procedures on the farm.
Use of ELISA serology for subclinical Salmonella infection surveillance
An increase in the number of laboratories using in-house Salmonella ELISA tests and the approval of commercial kits will make Salmonella serology more available to practitioners and producers. There has not been a large outbreak of human foodborne salmonellosis attributable to pork consumption in the United States. It remains to be seen if or when serologic surveillance is widely adopted in the United States. When reports regarding use and interpretation of Salmonella serological testing are examined, three facts are clear. First, serological testing is most useful on a herd level, to identify herds with elevated Salmonella exposure. Second, repeated testing of successive groups of pigs in a herd is important to gain an accurate assessment of the Salmonella status of the herd. Finally, serological testing should not be used to identify a pig or group of pigs as Salmonella-infected or Salmonella-free. In a herd with high seroprevalence, culture of pooled pen fecal samples may identify the serotypes present. Considerations in selecting a Salmonella ELISA include the ability of the test to detect indigenous serotypes, availability of the test, and cost per sample. Information on the "cutoff" value and an approximation of how the test generally compares with other tests used in the same geographic area are also important.
Experience with serological testing in other countries suggests that it be used for seroprevalence determination and ongoing monitoring to provide a means of establishing a baseline for the herd and identifying a change in seroprevalence. That is, the test should be able to determine whether intervention strategies are warranted and, if implemented, are effective. Serological testing forSalmonella serovars represents another tool in the management of on-farm food-safety procedures for swine producers and veterinarians.
Appendix
Danish monitoring for slaughter pig producing herds
To approximate Danish-type surveillance for a herd, first establish a baseline index. For 3 consecutive months, either collect serum samples from 10 to 20 animals close to slaughter, or obtain 10 to 20 meat juice samples from carcasses after slaughter. Assay the samples for Salmonella antibody with the DME27 or an equivalent ELISA.28-35 In the Danish sampling system, 60, 75, or 100 samples per year are collected from each herd, depending upon herd size. Calculate the seroprevalence of the group, i.e., the number of positive samples (as determined by the "cutoff value" of the specific test you are using) divided by the total number tested. Calculate a weighted average of the seroprevalence (weighted 0.2, 0.2, and 0.6, least to most recent test), which provides a figure roughly equivalent to the Danish Salmonella Index. If the weighted average is <40, the herd would be considered Level 1 (low seroprevalence). If it is 40 to 70, the herd is Level 2, and if it is >70, the herd is Level 3 (high seroprevalence). This procedure is described and an example of this calculation is provided by Alban et al.14 Danish control programs for Salmonella have recently been reviewed by Wegener et al36 and elimination and eradication procedures by Harris and Harris.37 Salmonella ELISAs should not be used to classify a particular group of animals as "infected" with Salmonella. An antibody response is a historical measure and does not mean the animal is still infected; lack of an antibody response does not mean the animal is free from infection.
Acknowledgements
The author would like to extend her sincere appreciation to the following individuals and granting agencies: Drs Dave Baum, Thomas Blaha, Eric Bush, Jan Dahl, Mike Daniels, Bill Christianson, Jeff Gray, D.L. Harris, N. Lee, Ann Letellier, Al Loynachan, Denis Matousek, Bent Nielsen, D. Nilubol, Bo Norby, L.L. Sorensen, Montserrat Torremorell, and Rick Tubbs; Chris Baum, Brad Chriswell, Matt Erdman, Kathy Ferris, Stephen Gaul, Ellen Martens, Jeanne Nugent, Stephanie Wedel, Yuhua Zhang, and Huiyan Zhao; and the Food Safety Consortium (US Department of Agriculture [USDA]- Cooperative State Research, Education, and Extension Service #20013421110276); the National Pork Board (National Pork Producers' Council #00-100); PIC, US; and the USDA (#00511109757).
References - refereed
2. Baggesen DL, Wegener HC, Bager F, Stege H, Christensen J. Herd prevalence of Salmonella enterica infection in Danish slaughter herds determined by microbiological testing. Prev Vet Med. 1996;26:201-213.
5. Christensen J, Gardner IA. Herd-level interpretation of test results from epidemiologic studies of animal diseases. Prev Vet Med. 2000;45:83-106.
8. Mousing J, Thode Jensen P, Halgaard, Bager F, Feld N, Nielsen B, Nielsen JP, Bech-Nielsen S. Nation-wide Salmonella enterica surveillance and control in Danish slaughter swine herds. Prev Vet Med. 1997;29:247-261.
14. Alban L, Stege H, Dahl J. The new classification system for slaughter-pig herds in the Danish Salmonella surveillance-and-control program. Prev Vet Med. 2002;53:133-146.
23. Charles SD, Abraham AS, Trigo ET, Jones GF, Settje TL. Reduced shedding and clinical signs of Salmonella Typhimurium in nursery pigs vaccinated with a Salmonella Choleraesuis vaccine. Swine Health Prod. 2000;8:107-112.
27. Nielsen B, Baggesen D, Bager F, Haugegaard J, Lind P. The serological response to Salmonella serovars Typhimurium and Infantis in experimentally infected pigs. The time course followed with an indirect anti-LPS ELISA and bacteriological examinations. Vet Micro. 1995;47:205-218.
29. Davies PR, Morrow WEM, Jones FT, Deen J, Fedorka-Cray PJ, Gray JT. Risk of shedding Salmonella organisms by market-age hogs in a barn with open-flush gutters. JAVMA. 1997;210:386-389.
32. Jauho ES, Boas U, Wiuff C, Wredstrom K, Pedersen B, Andresen LO, Heegaard PMH, Jakobsen MH. New technology for regiospecific covalent coupling of polysaccharide antigens in ELISA for serological detection. J Immunol Meth. 2000;242:133-143.
33. Wiuff C, Jauho ES, Stryhn H, Andresen LO, Thaulov K, Boas U, Jakobsen MH, Heegaard PMH. Evaluation of a novel enzyme-linked immunosorbent assay for detection of antibodies against Salmonella, employing a stable coating of lipopolysaccharide-derived antigens covalently attached to polystyrene microwells. J Vet Diagn Invest. 2000;12:130-135.
36. Wegener HC, Hald T, Wong DLF, Madsen M, Koragaard H, Bager F, Gerner-Smidt P, Molbak K. Salmonella control programs in Denmark. Emerg Inf Dis. 2003;9:774-780.
References - non refereed
1. Van der Heijden HMJF. First International ring trial of ELISAs for Salmonella-antibody detection in swine. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;481-491.
3. Erdman MM, Harris IT, Harris DL. Isolation of Salmonella using pooled pen feces from 37 U.S. swine farms. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;547-549.
4. Ferris KE, Flugrad BR, Timm JM, Ticer AE. Salmonella serotypes from animals and related sources reported during July 1999-June 2000. Proc USAHA Meet. Birmingham, Alabama. 2000;521-526.
6. Sorensen LL, Dahl J, Nielsen B. Correlation between Salmonella serology and results from bacteriological examinations of caecal contents, carcass swabs, pharyngeal swabs and caecal lymph nodes from Danish slaughter pigs. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;316-318.
7. Torremorell M, Turney-Harris I, Gramer M, Donovan T, Harris DL. Serologic monitoring for Salmonella in multi-site systems. Proc IPVS. Melbourne, Australia. 2000;208.
9. Baum DH. Vaccine and epidemiologic studies of Salmonella infections in swine. [PhD dissertation]. Ames, Iowa: Iowa State University; 1997.
10. Dahl J. The relationship between Salmonella-shedding and the Danish Salmonella-mix-ELISA on the pig level. Proc IPVS. Birmingham, England. 1998;279.
11. Funk JA, Harris IT, Gray JT, Davies PR. Comparison of traditional fecal culture, Danish mix-ELISA and SalAD for determination of Salmonella enterica prevalence in growing swine. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;515-517.
12. Gibson K, Ritter L, Blaha T, Carlson A, Szaszak A, Maes D, Grass J, Harris IT. Monitoring the dynamics of Salmonella prevalence in commercial swine herds. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;274-280.
13. Harris IT, Harris DL, Martens E. Serologic monitoring of U.S. pig herds for Salmonella. Proc IPVS. Melbourne, Australia. 2000;209.
15. Gray JT, Fedorka-Cray PJ. Long term survival and infectivity of Salmonella Choleraesuis. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;444-446.
16. Wingstrand A, Christensen G, Thomsen LK, Dahl J, Jensen BB. Reduction of subclinical Salmonella infection by feeding coarse ground feed and adding formic acid to water. Proc IPVS. Bologna, Italy. 1996;180.
17. Wingstrand A, Dahl J, Thomsen LK, Jorgensen L, Jensen BB. Influence of dietary administration of organic acids and increased feed structure on Salmonella Typhimurium infection in pigs. Proc 2nd Int Symp Epidemiol Cont Salmonella Pork. 1997;170-172.
18. Dahl J, Wingstrand A, Baggesen DL, Nielsen B, Thomsen LK. The effect of commercial organic acid preparations on seroprevalence and shedding of Salmonella in finishing pigs. Proc IPVS. Bologna, Italy. 1996;178.
19. Dahl J. The effect of feeding non-heat treated, non-pelleted feed compared to feeding pelleted, heat treated feed on the Salmonella-seroprevalence of finishing pigs. Proc IPVS. Birmingham, England. 1998;125.
20. Dahl J, Wingstrand A. The effect of dietary administration of an organic acid preparation on Salmonella seroprevalence. Proc IPVS. Birmingham, England. 1998;178.
21. Baum DH, Harris DL, Roof MB, Nielsen B, Holck JT, Polson DP, Baik J. Use of SC54 for the reduction of Salmonella in swine. Proc 2nd Int Symp Epidemiol Cont Salmonella Pork. 1997;215-218.
22. Charles SD, Trigo E, Settje T, Blaha TH, Gibson KJ, Frank RK. Evaluation of cross protection afforded by a cya (crp-cdt) Salmonella Choleraesuis commercial vaccine against Salmonella Typhimurium infection in pigs. Proc AASP. Indianapolis, Indiana. 2000;105-106.
24. Gibson KJ, Blaha TH, Frank RK, Charles SD, Trigo E. Investigation into the capability of a Salmonella Choleraesuis live vaccine to reduce the shedding of Salmonella Typhimurium in swine. Proc 3rd Int Symp Epidemiol Cont Salmonella Pork. 1999;302-304.
25. Kolb JR, Burkart K. Reduction of Salmonella species contamination of swine carcasses utilizing vaccination with an avirulent live Salmonella Choleraesuis vaccine (SC-54) at placement in grower/finisher. Proc IPVS. Birmingham, England. 1998;74.
26. Letellier A, Messier S, Lessard L, Quessy S. Host response to different treatments to reduce Salmonella infections in swine. Proc 3rd Int Symp Epidemiol Cont Salmonella Pork. 1999;317-320.
28. Gray JT, Fedorka-Cray PJ. Detection of swine exposed to Salmonella spp. Proc 3rd Int Symp Epidemiol Cont Salmonella Pork. 1999;46-50.
30. Letellier A, Quessy S. Determination of the Salmonella status of swine herds by the use of an ELISA test. Proc Conf Res Work Anim Dis. 2000: Abstract 42P.
31. Letellier A, Cote S, Surprenant C, Quessy S. Use of serology to evaluate the impact of clinical salmonellosis in swine on the herd status and on the contamination of pig carcasses from affected herds. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;319-321.
34. Gabert J, Schalch B, Greil B, Sperner B, Stolle A, Wever C, Kramer T. The use of a commercial test system (SalmotypeR-ELISA) for tracing antibodies to Salmonella in the serum of pigs. Proc 3rd Int Symp Epidemiol Cont Salmonella Pork. 1999;37-41.
35. Camitz A, Holmquist G, Ballagi A, Rodgers S. HerdChek Salmonella antibody ELISA for the serological monitoring of Salmonella infection in swine. Proc 4th Int Symp Epidemiol Cont Salmonella other Food Path Pork. 2001;505-508.
37. Harris DL, Harris IT. Salmonella elimination and reduction. Proc Int Symp Swine Dis Erad. 2001;51-59.
