| Brief communication | Peer reviewed |
Cite as: Jacques M, Grenier D, Labrie J, et al. Persistence of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 in bacterial biofilms. J Swine Health Prod. 2015;23(3):132–136.
Also available as a PDF.
SummaryThe aim of this pilot project was to investigate association of viruses with bacterial biofilms. Our preliminary data indicate that important viral pathogens of swine, namely, porcine reproductive and respiratory syndrome virus and porcine circovirus type 2, can associate with and persist within bacterial biofilms for several days. | ResumenLa meta de este proyecto piloto fue investigar la asociación de virus con biofilms bacterianos. Nuestra información preliminar indica que importantes patógenos virales porcinos, o sea, virus del síndrome reproductivo y respiratorio y circovirus porcino tipo 2, pueden asociarse y persistir dentro de los biofilms bacterianos por varios días. | ResuméLe but de ce projet pilote était d’examiner l’association de virus avec des biofilms bactériens. Nos données préliminaires indiquent que des virus pathogènes importants chez le porc, en particulier le virus du syndrome reproducteur et respiratoire porcin et le circovirus porcin de type 2, peuvent être associés et persister pendant plusieurs jours à l’intérieur de biofilms bactériens. |
Keywords: swine, porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, persistence, bacterial biofilms, PRRS, PCV
Search the AASV web site
for pages with similar keywords.
Received: May 8, 2014
Accepted: September 16, 2014
Bacterial biofilms are structured clusters of bacterial cells that are enclosed in a self-produced polymer matrix and attached to a surface.1,2 Biofilms protect bacteria and allow them to survive and thrive under hostile environmental conditions. Bacteria within a biofilm are usually more resistant to elimination by immune cells and to the action of antibiotics and disinfectants. The latter represents an important problem for the food industry.3
The biofilm polymer matrix might also be able to protect viruses. It has been reported4 that the largemouth bass virus can associate with biofilms produced by environmental strains of Pseudomonas, and consequently the virus is protected against certain chemical disinfectants. Moreover, biofilms in drinking-water distribution systems can become reservoirs for pathogens, including enteric viruses.5 We thus hypothesized that important viral pathogens of swine can associate with bacterial biofilms and persist for long periods in the environment of swine farms. The aims of this pilot project were to investigate the association of two important viral pathogens of swine, namely, porcine reproductive and respiratory syndrome virus (PRRSV; an enveloped virus) and porcine circovirus type 2 (PCV2; a non-enveloped virus) with bacterial biofilms, and to determine whether bacterial biofilms can protect PCV2 against disinfectants.
Materials and methods
Bacterial biofilms
A standard microtiter plate assay for biofilm formation that is routinely used in our laboratory, and which involves staining biofilms with crystal violet, was performed.6 First, biofilms of enteric bacterial pathogens (Escherichia coli strain ECL 17608 or Salmonella Typhimurium strain ATCC 14028) and respiratory bacterial pathogens (Actinobacillus pleuropneumoniae serotype 1 strain 719 or Streptococcus suis serotype 2 strain 735 and non-typeable strain 1097925) were established in vitro following an incubation of 24 to 72 hours. The growth conditions enabling optimal biofilm formation for these bacterial strains have been already determined in our laboratory.7-9 In some experiments, biofilms were visualized by confocal laser scanning microscopy.7
Persistence of PRRSV and PCV2 in bacterial biofilms
A defined amount of a virus preparation (PRRSV genotype 2 reference strain IAF-Klop, 103.0 median tissue culture infective doses [TCID50] per well; or PCV2b strain FMV06-0732, 104.5 TCID50 per well) was added to the culture of each of the five named bacterial pathogens and incubated in a standard microtiter plate assay for biofilm formation. The persistence of the viral genome was monitored for up to 3 days in both the supernatant (ie, the liquid phase above the biofilm) and the biofilm attached at the bottom of the well. Virus-specific quantitative polymerase chain reaction (qPCR) and reverse-transcriptase qPCR (RT-qPCR) assays were performed according to standardized protocols used by the Diagnostic Services (Faculté de médecine vétérinaire, Université de Montréal), namely, an in-house assay for PCV210 and a commercial kit for PRRSV (EZ-PRRSV MPX 4.0; Tetracore, Rockville, Maryland). The qPCR and RT-qPCR results were expressed in TCID50 per mL after arithmetically comparing them to standard curves previously established with infectious PRRSV and PCV2 titrated in cell cultures. Since a positive qPCR result does not necessarily correlate with infectious potential, viral infectious titers were also determined using specific permissive cell culture models: MARC-145 for PRRSV11 and NPTr12 for PCV2 (a new cell line permissive to PCV2; unpublished data). Bacterial cells were removed by filtration on a 0.22-μm pore size membrane (UFC30GVOS; EMD Millipore, Mississauga, Ontario, Canada) before titration. The amount of infectious virus was calculated from a 96-well microplate of infected cells by the Kärber method, and the results were expressed in TCID50 per mL.10 The survival of the virus within a biofilm was arithmetically compared to the survival of an equal amount of virus in a microtiter plate well in the absence of a biofilm.
PCV2 susceptibility to disinfectants in the presence of bacterial biofilms
In addition to evaluating viral persistence within bacterial biofilms, this study also determined whether bacterial biofilms can protect PCV2 (a non-enveloped virus known to be more resistant than PRRSV, an enveloped virus) against disinfectants. Porcine circovirus type 2 virions, in the presence or absence of A pleuropneumoniae biofilms, were exposed for 30 minutes to several classes of disinfectants routinely used on farms at the concentrations recommended by the manufacturers (1% acid peroxygen; Virkon, Vétoquinol, Lavaltrie, Quebec, Canada; and 1% quaternary ammonium-glutaraldehyde; Aseptol 2000, S.E.C. Repro Inc, Ange-Gardien, Quebec, Canada). A virus-specific qPCR assay could not be used in these experiments, since a positive qPCR does not correlate with infectious potential. The infectious viral titers were thus determined using the appropriate cell line as described. To ensure that residual disinfectant did not interfere with the assay, excess disinfectant was removed by ultracentrifugation at 100,000g for 1 hour, and the virus pellet was resuspended in water to the initial volume. The viability of the bacterial cells within the biofilm was also evaluated after exposure to the disinfectants using the CellTiter-Blue cell viability assay (Promega, Madison, Wisconsin).
Results
Persistence of PRRSV and PCV2 in bacterial biofilms
An example of a bacterial biofilm formation is shown in Figure 1. Results of monitoring the presence of PRRSV for up to 3 days using a virus-specific qPCR assay are presented in Table 1. A small proportion of the viral inoculum persisted in the biofilms for the duration of the experiment; this amount was considered too small, however, to attempt quantification of infectious viruses by titration on the MARC-145 cell line. For example, the amount of PRRSV recovered from the E coli biofilm was 16 to 44 TCID50 per mL for all time points tested, compared to a much greater amount in positive control wells (4831 to 5880 TCID50 per mL). Results of monitoring for the presence of PCV2b using the qPCR assay showed, again, that a portion of the viral inoculum persisted in the biofilms for the duration of the experiment (Table 2). These viruses were infectious when inoculated onto NPTr cells (data not shown).
Figure 1: Confocal laser scanning microscopic image of a biofilm of Actinobacillus pleuropneumoniae, an important bacterial swine pathogen. This is a top view of a biofilm formed at the bottom of a microtiter plate well after an incubation of 5 hours. The biofilm was stained with wheat germ agglutinin (WGA)-Oregon Green 488, a lectin that binds to poly-N-acetylglucosamine in the biofilm matrix.
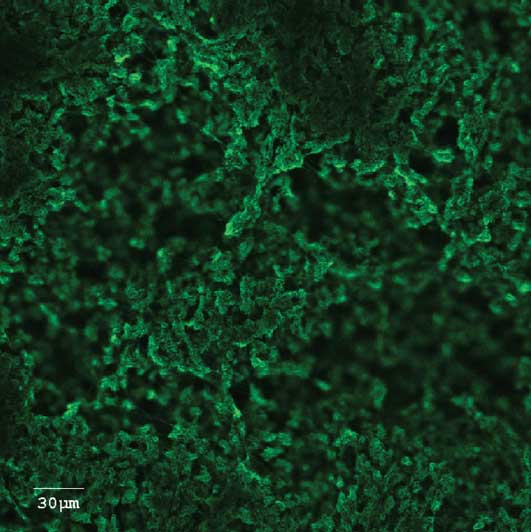
Table 1: Results of PCR testing for PRRSV in biofilms of Actinobacillus pleuropneumoniae, Escherichia coli, Salmonella, and Streptococcus suis
| TCID50 of PRRSV/mL | ||||
|---|---|---|---|---|
| 24 hours | 48 hours | 72 hours | ||
| A pleuropneumoniae | Supernatant | 359 | 2109 | 858 |
| Biofilm | 27 | 14 | 4 | |
| Positive control† | 2427 | 4024 | 685 | |
| Negative control† | 0 | 0 | 0 | |
| E coli | Supernatant | 5758 | 4409 | 3214 |
| Biofilm | 16 | 14 | 44 | |
| Positive control† | 4831 | 5292 | 5880 | |
| Negative control† | 0 | 0 | 0 | |
| Salmonella | Supernatant | 252 | 344 | 219 |
| Biofilm | 21 | 28 | 53 | |
| Positive control† | 1464 | 3329 | 3622 | |
| Negative control† | 0 | 0 | 0 | |
| S suis | Supernatant | 2487 | 2828 | 2638 |
| 735‡ | Biofilm | 106 | 67 | 276 |
| Positive control† | 5035 | 3713 | 3031 | |
| Negative control† | 0 | 0 | 0 | |
| S suis | Supernatant | 4574 | 4110 | 3654 |
| NT1097925§ | Biofilm | 377 | 258 | 256 |
| Positive control† | 5035 | 3713 | 3031 | |
| Negative control† | 0 | 0 | 0 | |
* For each organism, the virus suspension was added to a bacterial culture and incubated in a standard microtiter plate assay for biofilm formation. Testing for PRRSV was conducted daily, using a commercial PCR kit (EZ-PRRSV MPX 4.0; Tetracore, Rockville, Maryland), for up to 3 days in the supernatant (liquid phase above the biofilm) and in the biofilm attached to the plastic surface.
† Positive control, no bacteria, virus only; negative control, no virus, bacteria only.
‡ This strain produces a biofilm in the presence of fibrinogen.8
§ This strain does not require fibrinogen to produce a biofilm.9
PCR = polymerase chain reaction; PRRSV = porcine reproductive and respiratory syndrome virus; TCID50 = median tissue culture infectious dose.
Table 2: Results of testing for PCV2b in Actinobacillus pleuropneumoniae, Escherichia coli, Salmonella, and Streptococcus suis biofilms by virus-specific PCR (expressed as TCID50/mL)*
| TCID50 of PCV2b/mL | ||
|---|---|---|
| A pleuropneumoniae | Supernatant | 7.21 × 105 |
| Biofilm | 5.63 × 104 | |
| Positive control† | 9.77 × 105 | |
| Negative control† | 0 | |
| E coli | Supernatant | 4.99 × 105 |
| Biofilm | 5.89 × 104 | |
| Positive control† | 9.68 × 105 | |
| Negative control† | 0 | |
| Salmonella | Supernatant | 1.78 × 104 |
| Biofilm | 1.74 × 104 | |
| Positive control† | 7.90 × 105 | |
| Negative control† | 0 | |
| S suis | Supernatant | 7.71 × 104 |
| 735‡ | Biofilm | 3.57 × 104 |
| Positive control† | 3.48 × 105 | |
| Negative control† | 0 | |
| S suis | Supernatant | 3.41 × 105 |
| NT1097925§ | Biofilm | 5.15 × 104 |
| Positive control† | 3.48 × 105 | |
| Negative control† | 0 |
* The virus suspension was added to a bacterial culture and incubated in a standard microtiter plate assay for biofilm formation for 24 hours for A pleuropneumoniae, E coli, and S suis, and for 48 hours for Salmonella in order to achieve optimal biofilm formation. The presence of the virus was determined for up to 2 days in the supernatant (liquid phase above the biofilm) and in the biofilm attached to the plastic surface.
† Positive control: no bacteria, virus only; negative control, no virus, bacteria only.
‡ This strain produces a biofilm in the presence of fibrinogen.8
§ This strain does not require fibrinogen to produce a biofilm.9
PCV2b = porcine circovirus type 2b; PCR = polymerase chain reaction; TCID50 = median tissue culture infectious dose.
PCV2 susceptibility to disinfectants in the presence of bacterial biofilms
In the second set of experiments, a defined amount of PCV2b virus strain FMV06-0732 was added to a culture of A pleuropneumoniae, assayed for biofilm formation, and subsequently treated with disinfectants. Results showed that PCV2b titers were lower in the presence of each disinfectant than in the negative control (ie, wells without disinfectants) and that the efficacy of the disinfectants against PCV2b was only slightly lower in the presence of the A pleuropneumoniae biofilm than in the control (Table 3).
Table 3: Detection of PCV2b infectious viral particles (TCID50/mL) in an Actinobacillus pleuropneumoniae biofilm*
| TCID50 of PCV2b/mL (log difference from control) | |||
|---|---|---|---|
| Control | Quaternary ammonium-glutaraldehyde | Acid peroxygen | |
| Wells without biofilm | 104.00† | 102.90† (1.10) | 102.80† (1.20) |
| Wells with A pleuropneumoniae biofilm | 102.60‡ | 101.75‡ (0.85) | 101.90‡ (0.70) |
* The virus suspension (PCV2b virus strain FMV06-0732) was added to the bacterial culture and incubated in a standard microtiter plate assay for biofilm formation. Porcine circovirus type 2 virions, in the presence or absence of biofilms, were then exposed to acid peroxygen and quaternary ammonium-glutaraldehyde disinfectants at 1% each and for 30 minutes of exposure. Control wells contained no disinfectant. Infectious viral titers were determined using the NPTr cell line. The numbers of infectious viruses in the biofilm attached to the plastic surface were arithmetically greater than the numbers in wells without a biofilm.
† Number of infectious viruses present in the liquid suspension.
‡ Number of infectious viruses present in the biofilm phase only.
PCV2b = porcine circovirus type 2b; TCID50 = median tissue culture infectious dose.
Results of experiments using the CellTiter-Blue cell viability assay to test the antibacterial efficacy of both disinfectants against A pleuropneumoniae biofilms clearly indicated that both disinfectants were effective to decrease the metabolic activity of A pleuropneumoniae under the study conditions.Overall, metabolic activity was lower by 90% to 100% in the biofilm samples treated with disinfectants than in the non-treated samples (data not shown), suggesting that large numbers of bacterial cells had died. It is important to note that the disinfectants killed the bacterial cells but did not remove the biofilms.
Discussion
The effects of biofilms of enteric (E coli or Salmonella) or respiratory (A pleuropneumoniae or S suis) bacterial pathogens on PRRSV and PCV2, two of the most important viruses in the swine industry, were studied. Several control experiments were conducted prior to initiating this pilot project to evaluate, for example, methods for virus recovery from a bacterial biofilm, or to ensure that no traces of disinfectant were left that would affect the cell lines used for viral titration. Overall, our results indicate that a small portion of the viral inoculum persisted in the biofilms for the duration of the experiments, as first determined by qPCR. The amount of PRRSV was too small to attempt infectious virus quantification by titration on MARC-145 cell lines. However, the amount of PCV2 was greater, and viral titration on NPTr cells was performed, confirming that a proportion of the PCV2 inoculum persisted in the biofilms and remained infectious. A recent publication13 indicated that binding of an enteric virus (poliovirus) to bacterial polysaccharides stabilizes the virions and may offer a selective advantage by enhancing environmental stability.
Although the amounts of virus persisting in bacterial biofilms were relatively low for one of the tested viruses (namely, PRRSV), there was a possibility that once incorporated in a biofilm, these viruses would be protected from disinfectants. Our preliminary results indicate that bacterial biofilms seem to only slightly reduce the efficacy of disinfectants, which nevertheless remain effective against the virus tested.
To the best of our knowledge, this is the first description of the persistence of two important swine viral pathogens, PRRSV and PCV2b, within bacterial biofilms. This pilot project generated preliminary data important for the swine industry in a new area that certainly deserves to be investigated in more detail. It would be relevant to perform similar experiments with other important swine viruses such as the porcine epidemic diarrhea virus or the porcine deltacoronavirus.
Implications
• PRRSV and PCV2 can associate with bacterial biofilms that are known to be present inside the infected host or in the farm’s environment.
• PRRSV and PCV2 can persist within Gram-positive and Gram-negative bacterial biofilms for several days.
• Under the conditions of this study, the efficacy of acid peroxygen and quaternary ammonium-glutaraldehyde commercial disinfectants against PCV2 may be only slightly reduced by the presence of a bacterial biofilm.
Acknowledgements
This research was supported by grants by the Canadian Swine Health Board (to MJ and CAG) and the Swine and Poultry Infectious Diseases Research Centre (to MJ, DG, and CAG). We thank Drs J. M. Fairbrother, M. Gottschalk, and A. Letellier for bacterial strains.
Conflict of interest
None reported.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
1. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322.
2. Jacques M, Aragon V, Tremblay YDN. Biofilm formation in bacterial pathogens of veterinary importance. Anim Health Res Rev. 2010;11:97–121.
3. Van Houdt R, Michiels CW. Biofilm formation and the food industry, a focus on the bacterial outer surface. J Appl Microbiol. 2010;109:1117–1131.
4. Nath S, Aron GM, Southard GM, McLean RJ. Potential for largemouth bass virus to associate with and gain protection from bacterial biofilms. J Aquat Anim Health. 2010;22:95–101.
5. Wingender J, Flemming HC. Biofilms in drinking water and their role as reservoir for pathogens. Int J Hyg Environ Health. 2011;214:417–423.
6. Labrie J, Pelletier-Jacques G, Deslandes V, Ramjeet M, Auger E, Nash JH, Jacques M. Effects of growth conditions on biofilm formation by Actinobacillus pleuropneumoniae. Vet Res. 2010;41:03. doi:10.1051/vetres/2009051.
7. Wu C, Labrie J, Tremblay YD, Haine D, Mourez M, Jacques M. Zinc as an agent for the prevention of biofilm formation by pathogenic bacteria. J Appl Microbiol. 2013;115:30–40.
8. Bonifait L, Grignon L, Grenier D. Fibrinogen induces biofilm formation in Streptococcus suis and enhances its antibiotic resistance. Appl Environ Microbiol. 2008;74:4969–4972.
9. Bonifait L, Gottschalk M, Grenier D. Cell surface characteristics of nontypeable isolates of Streptococcus suis. FEMS Microbiol Lett. 2010;311:160–166.
10. Gagnon CA, del Castillo JR, Music N, Fontaine G, Harel J, Tremblay D. Development and use of a multiplex real-time quantitative polymerase chain reaction assay for detection and differentiation of Porcine circovirus-2 genotypes 2a and 2b in an epidemiological survey. J Vet Diagn Invest. 2008;20:545–558.
11. Lévesque C, Provost C, Labrie J, Hernandez Reyes Y, Burciaga Nava JA, Gagnon CA, Jacques M. Actinobacillus pleuropneumoniae possesses an antiviral activity against porcine reproductive and respiratory syndrome virus. PLoS One. 2014;9:e98434.
12. Ferrari M, Scalvini A, Losio MN, Corradi A, Soncini M, Bignotti E, Milanesi E, Ajmone-Marsan P, Barlati S, Bellotti D, Tonelli M. Establishment and characterization of two new pig cell lines for use in virological diagnostic laboratories. J Virol Methods. 2003;107:205–212.
13. Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15:36–46.