| Original research | Peer reviewed |
Cite as: Petrovan V, Fang Y, Rowland RRR. Presence of Senecavirus A in pork sold in the United States. J Swine Health Prod. 2019;27(2):87-91.
Also available as a PDF.
SummaryObjective: To estimate the prevalence and concentration of Senecavirus A (SVA) in meat sold at retail. Materials and methods: A total of 190 meat samples derived from 25 processing locations in 13 states were purchased through retail sources. The presence of virus in samples of muscle obtained from each package was assessed by polymerase chain reaction (PCR) amplification of SVA nucleic acid. A standard curve was constructed to estimate the concentration of viable virus in PCR-positive samples. Results: Two of the 190 meat samples (1.1%) were positive for SVA nucleic acid, but negative for virus by virus isolation. The amount of virus in the PCR-positive samples was estimated to be less than 14 virions/g of muscle. Implications: The low prevalence of SVA in the 190 retail-meat samples analyzed in this study, combined with a low concentration of SVA nucleic acid in the two SVA-positive samples, suggest a low risk for transmitting SVA through retail meat. | ResumenObjetivo: Estimar la prevalencia y concentración del Senecavirus A (SVA por sus siglas en inglés) en carne vendida al menudeo. Materiales y métodos: Un total de 190 muestras de carne procedente de 25 sitios de procesamiento en 13 estados fueron compradas a través de fuentes de venta al menudeo. La presencia del virus en muestras de músculo obtenidas de cada paquete fue evaluada por medio de la amplificación de la reacción en cadena de polimerasa (PCR por sus siglas en inglés) del ácido nucleico del SVA. Se elaboró una curva estándar para estimar la concentración de virus viable en muestras positivas al PCR. Resultados: Dos de las 190 muestras de carne (1.1%) resultaron positivas al ácido nucleico del SVA, pero resultaron negativas al virus por medio de aislamiento viral. Se estimó que la cantidad de virus en las muestras positivas al PCR era menos de 14 viriones/g de músculo. Implicaciones: La baja prevalencia de SVA en las 190 muestras de carne vendida al menudeo analizadas en este estudio, en conjunto con la baja concentración de ácido nucleico SVA en las dos muestras positivas al SVA, sugieren un bajo riesgo de transmisión del SVA por medio de la carne vendida al menudeo. | ResuméObjectif: Estimer la prévalence et la concentration de Senecavirus A (SVA) dans la viande vendue au détail. Matériels et méthodes: Un total de 190 échantillons de viande obtenu de 25 usines de transformation dans 13 états furent achetés dans des magasins de vente au détail. La présence du virus dans des échantillons de muscle obtenus de chaque emballage était déterminée par réaction d’amplification en chaine par la polymérase (PCR) de l’acide nucléique du SVA. Une courbe standard fut élaborée pour estimer la concentration de virus viables dans les échantillons positifs par PCR. Résultats: Deux des 190 échantillons de viande (1.1 %) étaient positifs pour l’acide nucléique de SVA, mais négatifs pour l’isolement viral. On estima à moins de 14 virions/g de muscle la quantité de virus dans les échantillons positifs par PCR. Implications: La faible prévalence de SVA dans les 190 échantillons de viande analysés dans la présente étude, ainsi que la faible concentration d’acide nucléique de SVA dans les deux échantillons positifs, suggèrent un faible risque de transmission de SVA via la viande vendue au détail. |
Keywords: swine, Senecavirus A, SVA, SVA in meat
Search the AASV web site
for pages with similar keywords.
Received: May 25, 2018
Accepted: September 26, 2018
Senecavirus A (SVA), also known as Seneca Valley virus, is a single-strand, non-enveloped RNA virus belonging to the genus Senecavirus, family Picornaviridae.1,2 Important foreign animal disease (FAD) viruses in this family include foot-and-mouth disease virus (FMDV) and swine vesicular disease virus (SVDV). Similarities with FMDV in terms of physiochemical properties make SVA a suitable surrogate for understanding the environmental stability of FMDV.3 The key clinical sign associated with FMDV or SVDV infection of pigs is the formation of vesicular lesions on the snout and feet.4 In 2004, an outbreak of idiopathic vesicular disease occurred in a farrow-to-finish farm in Indiana.5 Extensive analysis showed that pigs were negative for FMDV, SVDV, and other agents associated with vesicular lesions. The infected pigs eventually recovered, but vesicular disease signs reappeared in the herd. Pasma et al6 identified SVA as the source of the vesicular disease syndrome. The virus possessed a nucleic acid sequence closely related to a virus originally isolated in Brazil.7-9 Experimental infection studies confirmed the ability of SVA to cause vesicular lesions.10 However, it should be noted that SVA can be present in pigs without signs of overt clinical disease.
Perhaps the greatest impacts of SVA infection on swine production are the consequences of finding vesicular lesions. Because vesicular lesions are associated with FMDV and SVDV, the appearance of lesions results in herd closure followed by time-consuming FAD investigations involving local, state and federal authorities.
It is well established that pig meat is a potential vector for introducing disease into naïve populations.11 For meat to be a risk for infection, the virus must be present in a sufficiently high quantity to deliver an infectious oral dose to a susceptible animal. A likely mechanism for disease introduction is via uncooked meat scraps or spoiled meat discarded as garbage. Infection could occur through the consumption of the discarded meat by feral pigs, which then could come into contact with domestic pigs. Another route for introduction is through the incorporation of meat scraps into unprocessed swill fed to backyard pigs.
The purpose of this study was to investigate the potential risk of introducing SVA through meat by estimating the prevalence and concentration of SVA nucleic acid in muscle meats purchased at retail. Because the time from slaughter to purchase of a pork product in a retail store within the United States is similar to the time needed to transport pork to another country via legal trade, and because the steps and processes involved are also comparable, the condition and age of US pork products for sale at retail in the United States and other countries is also similar. Determining the current prevalence of SVA in retail products in the United States can subsequently aid in risk analyses surrounding SVA in pork.
Materials and methods
Collection and processing of retail meat samples
There are a limited number of studies that provide an estimation of the prevalence of SVA infection at the time of slaughter. Based on Hause et al,9 we selected a prevalence of 1% to 5%, which is a conservative estimate. We would expect that the assay of 200 meat samples would yield 2 to 9 positive results. Of the 200 samples, 190 were successfully assayed. The ten samples not assayed consisted of products containing chopped or ground pork or were subjected to processing (ie, smoking, curing, or marinating). Sampling bias was avoided by selecting a variety of cuts from six different retail supermarket chains located in Manhattan (five individual stores), Junction City (three stores), and Kansas City, Kansas (one store) and Alexandria, Virginia (one store). Sampling occurred on 15 days over a 2-month period between February 28, 2017 and April 30, 2017. Meat was collected from one to five stores per day. Based on establishment code numbers (ESTN), the origin of each package was traced to 1 of 25 meat processing locations in 13 states (Table 1). Effort was made to select packages that possessed unique ESTN to ensure that samples were collected from the greatest number of meat processing facilities. Ten facilities were in the Midwest: Colorado, Iowa, Illinois, Kentucky, Minnesota, Missouri, Nebraska, South Dakota, Texas, and Wisconsin. The remaining processing facilities were in California, North Carolina, and Virginia. Muscle meat samples with and without bone were analyzed including chops (41 samples), loins (61 samples), ribs (53 samples), roasts (9 samples), shoulders (18 samples), and other (8 samples). Less common cuts sampled, such as feet, carnitas (chopped meat), neck bones, and cutlets, were identified as other. Ground products, such as ground pork and fresh sausage, were not part of this study, primarily because these products contain more than muscle meat and are generally not exported.
Table 1: Sampled cuts of meat and the processing establishment location from which they originated
| Cut of meat | |||||||
|---|---|---|---|---|---|---|---|
| State (No.*) | chop | loin | rib | roast | shoulder | other† | Total |
| CA (1) | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| CO (1) | 19 | 3 | 7 | 0 | 3 | 1 | 33 |
| IA (5) | 0 | 3 | 7 | 0 | 0 | 0 | 10 |
| IL (2) | 0 | 8 | 3 | 0 | 0 | 0 | 11 |
| KY (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| MN (5) | 0 | 12 | 9 | 1 | 0 | 0 | 22 |
| MO (2) | 8 | 4 | 6 | 3 | 0 | 0 | 21 |
| NC (2) | 0 | 0 | 4 | 0 | 0 | 0 | 4 |
| NE (2) | 3 | 16 | 8 | 0 | 8 | 1 | 36 |
| SD (1) | 0 | 10 | 4 | 5 | 4 | 0 | 23 |
| TX (1) | 10 | 2 | 4 | 0 | 2 | 6 | 24 |
| VA (1) | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
| WI (1) | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| Total | 41 | 61 | 53 | 9 | 18 | 8 | 190 |
* Number of processing establishments in each state represented in the sample.
† Less common cuts sampled, such as feet, carnitas (chopped meat), neck bones, and cutlets, were identified as other. Ground products, such as ground pork and fresh sausage, were not part of this study.
Refrigerated packages of meat were purchased at retail outlets and remained refrigerated until sample collection. Prior to sampling each package, all work surfaces and utensils were cleaned with 2% sodium hypochlorite (bleach) and thoroughly rinsed. Parchment paper was placed on the cleaned cutting surface. The exterior of each package was wiped down with bleach, assigned a unique identifier, and photographed to provide a record of product information and ESTN identifying the processing location. While wearing disposable gloves, a decontaminated knife or scissors was used to open the meat package. Using a fresh, decontaminated knife, a 5 to 10 g sample was excised, immediately placed in a plastic bag, and stored at -20°C until further processing to isolate RNA, typically within 48 hours. Between each sample collection, all utensils were soaked in bleach and thoroughly rinsed, surfaces were cleaned with bleach, and disposable gloves and surface parchment paper replaced.
RNA isolation and polymerase chain reaction of SVA nucleic acid
For isolation of total RNA, a 200 mg sample of muscle was placed in a GentleMACS M Tube. Four milliliters of TRIzol Reagent (Invitrogen, Waltham, Massachusetts) was added and the sample homogenized on a GentleMACS (Miltenyi Biotec, Auburn, California) for 84 seconds. Insoluble material was removed by centrifugation at 10,000g for 5 minutes. A 1-mL sample of supernatant was divided between two 1.5-mL microcentrifuge tubes and 0.5 mL of ethanol added to each tube. The RNA was isolated using a Direct-zol RNA MiniPrep Kit (R2052, Zymo Research, Irvine, California) according to the kit instructions. The RNA was eluted in a final volume of 50 μL of nuclease-free water and stored at -80°C. Polymerase chain reaction (PCR) was performed using the EZ-SVA Real Time RT-PCR detection kit (Tetracore, Rockville, Maryland). Briefly, a 25 μL reaction was carried out using 7 μL of extracted RNA and all steps performed according to the manufacturer’s instructions. Reverse transcription and amplification were performed on a CFX96 C1000 Thermal Cycler (BioRad, Hercules, California) under the following conditions: reverse transcription at 48°C for 15 minutes, initial denaturation at 95°C for 2 minutes, followed by 45 cycles of 95°C for 5 seconds and 60°C for 40 seconds. The high specificity of the commercial assay is based on the unique sequence of the primers specific for the SVA genomic sequence along with optimal PCR conditions used for amplification. This assay does not cross-react with other swine viruses. In terms of false-positive rates, the manufacture recommends that cycle threshold (Ct) values between 38 and 40 be retested.
Preparation of SVA standard curve
The sensitivity of the assay was determined by preparing a standard curve utilizing the SVA laboratory strain, KS15-01, which was originally isolated from a pig nasal swab sample by the Kansas Veterinary Diagnostic Laboratory. Virus was propagated, and the concentration measured on PK-15 cells as described previously.10 A standard curve for estimating the concentration of virus in meat samples was prepared by spiking 3.16 × 107 median tissue culture infective dose (TCID50) of virus into a 200 mg ground meat sample. The RNA was isolated from the SVA-spiked meat sample and further diluted to achieve a range of concentrations between 101 and 106 TCID50/g of tissue. The standard curve was plotted as log10 TCID50/g versus Ct value. For PCR, the standard curve and unknown meat samples were run on the same 96 well plate.
Results
Ten SVA standard curves were independently generated over the course of the study (Figure 1). The results showed a linear relationship between the Ct value and log virus concentration. The dilution containing approximately 10 TCID50 of virus approached a Ct of 40, which is the negative cutoff for the Tetracore PCR assay.
Figure 1: Generation of the SVA standard curve. Mean and standard deviation for 10 standard curves generated over the course of the study is shown along with the positive-negative cutoff for the Tetracore (Rockville, Maryland) SVA PCR assay, represented by the dashed line. SVA = Senecavirus A; PCR = polymerase chain reaction; Ct = cycle threshold, TCID50 = median tissue culture infective dose.
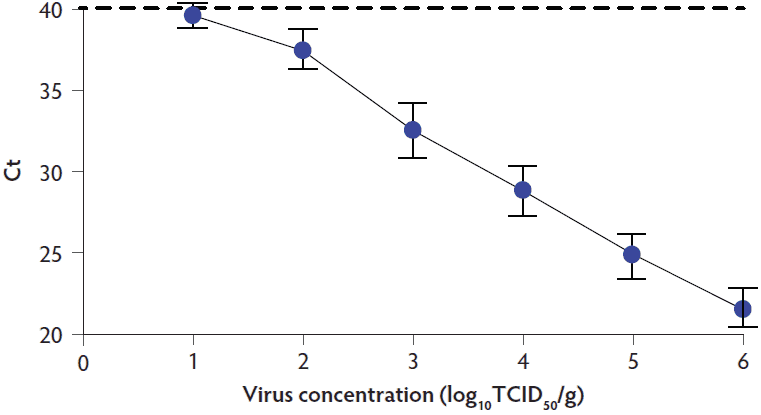
Of the 190 meat samples assayed, only 6 contained a sample of muscle that tested PCR-positive for SVA RNA (Ct value < 40; Table 2). Sample number 1033 and 1076 had Ct values of 36.6 and 37.0, respectively. Based on the standard curve, the estimated virus concentrations for the two positive samples were 1.4 log10 and 1.3 log10 TCID50/g, respectively. The four remaining samples possessed Ct values ranging from 38.4 to 39.2, which were considered borderline positive results. Samples, 1012A, 1022E and 1027G, were from packages that contained multiple cuts of meat. The remaining cuts of meat in each package were assayed and found to be PCR-negative (Ct > 40). The four suspect samples, 1012A, 1022E, 1027G, and 1029E, were subjected to PCR a second time and produced Ct values > 40. Together, these data show that only 2 of the 190 meat samples possessed detectable quantities of SVA nucleic acid. All PCR-positive samples were negative for the presence of viable virus by virus isolation on PK-15 cells.
Table 2: Packages of meat with PCR-positive samples for SVA RNA*
| Sample No. | Description | Cuts/pkg | Sample ID | Ct | Log10 TCID50/g |
|---|---|---|---|---|---|
| 1012 | Boneless sirloin chops | 4 | A | 38.4† | 1.0 |
| B | > 40 | < 1.0 | |||
| C | > 40 | < 1.0 | |||
| D | > 40 | < 1.0 | |||
| 1022 | Boneless chops | 7 | A | > 40 | < 1.0 |
| B | > 40 | < 1.0 | |||
| C | > 40 | < 1.0 | |||
| D | > 40 | < 1.0 | |||
| E | 38.8† | < 1.0 | |||
| F | > 40 | < 1.0 | |||
| G | > 40 | < 1.0 | |||
| 1027 | Boneless ribs | 7 | A | > 40 | < 1.0 |
| B | > 40 | < 1.0 | |||
| C | > 40 | < 1.0 | |||
| D | > 40 | < 1.0 | |||
| E | > 40 | < 1.0 | |||
| F | > 40 | < 1.0 | |||
| G | 39.2† | < 1.0 | |||
| 1029 | Boneless butt roast | 1 | A | 38.7† | < 1.0 |
| 1033 | Sparerib | 1 | A | 36.6 | 1.4 |
| 1076 | Loin | 1 | A | 37.0 | 1.3 |
* Samples with CT values < 40 were considered PCR-positive for SVA RNA.
† Samples were PCR-negative (Ct > 40) when assayed a second time.
PCR = polymerase chain reaction; SVA = Senecavirus A; pkg = package; ID = identification; Ct = cycle threshold; TCID50 = median tissue culture infective dose.
Discussion
Several factors are important for estimating the risk for the introduction of SVA through the exposure of pigs to retail pork products.11 The first consideration is the prevalence of SVA in the general US pig population. Based on PCR amplification of 2033 oral fluid samples from 25 states, Hause et al9 provided an estimated prevalence of SVA in the United States at about 1%. In a different study, incorporating the serological analysis of 5957 samples collected in 2016, seroprevalence was estimated at 28.95%.12 These later data represent pigs that are actively infected as well as pigs that were infected and subsequently cleared the virus. In the present study, 2 of 190 retail meat samples were found to be positive for SVA nucleic acid, supporting Hause’s estimate of 1% prevalence.9
A second consideration for transmission is the amount of virus present in meat. There are no published studies measuring the concentration of SVA in muscle. However, SVA nucleic acid can be detected in muscle tissue from heart and tongue of infected pigs.13 In the present study, the highest concentration of virus observed was estimated to be 1.5 log10 of virus/g of muscle, which was found in only 1 of 190 meat samples. The small amount of virus in the PCR-positive meat sample was supported by the negative results for virus isolation on PK-15 cells. Since there are no data on SVA concentration in different muscle meats, a wide variety of meats were tested, including both bone-in (n = 80) and boneless (n = 110) cuts.
A third factor related to the risk of transmission is the minimum infectious dose of SVA required to infect a pig when consuming meat. While there are no data for SVA, data are available for FMDV and SVDV. For example, Fukai et al14 showed that pigs given 106 virions of FMDV by direct oral administration resulted in three of three pigs becoming infected, whereas only one of three pigs were infected when administered 103 virions. However, Yamada et al15 failed to infect six pigs given an oral dose of 103 TCID50 of FMDV. For SVDV, the direct instillation of 5.3 log10 plaque forming units (pfu) in the mouth of pigs did not result in infection, while increasing the amount of virus to 6.8 log10 pfu resulted in three of six pigs becoming infected.16 If similar to SVDV and FMDV, the highest detected concentration of SVA in the present study (1.5 log10/g) would represent a negligible risk for transmission via the consumption of muscle meat by pigs.
In summary, the low prevalence of SVA combined with the low concentration of virus in positive meat samples, indicates a negligible risk for the transmission of SVA through the consumption of muscle meats sold at retail. Based on the study design, these results only apply to the United States. However, these results do also indicate a negligible risk for transmitting SVA from the United States through legal trade in pork.
Implications
- The low prevalence of SVA in the 190 retail-meat samples analyzed in this study, combined with a low concentration of SVA nucleic acid in the two SVA-positive samples, suggest a low risk for transmitting SVA through retail meat.
Acknowledgements
This work was supported by a grant from the National Pork Producers Council. A thank you from the authors to Carol Wyatt, retired professor, for reviewing the manuscript.
Conflict of interest
None reported.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
1. Adams MJ, Lefkowitz EJ, King AM, Bamford DH, Breitbart M, Davison AJ, Ghabrial SA, Gorbalenya AE, Knowles NJ, Krell P, Lavigne R, Prangishvili D, Sanfacon H, Siddell SG, Simmonds P, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2015;160:1837-1850.
2. Hales LM, Knowles NJ, Reddy PS, Xu L, Hay C, Hallenbeck PL. Complete genome sequence analysis of Seneca Valley virus-001 a novel oncolytic picornavirus. J Gen Virol. 2008;89:1265-1275.
3. Dee SA, Bauermann FV, Niederwerder MC, Singrey A, Clement T, de Lima M, Long C, Patterson G, Sheahan MA, Stoian AMM, Petrovan V, Jones CK, De Jong J, Ji J, Spronk GD, Minion L, Christopher-Hennings J, Zimmerman JJ, Rowland RRR, Nelson E, Sundberg P, Diel DG. Survival of viral pathogens in animal feed ingredients under transboundary shipping models. PLoS One. 2018;20:e0194509. doi:10.1371/journal.pone.0194509
4. Joshi LR, Fernandes MH, Clement T, Lawson S, Pillatzki A, Resende TP, Vannucci FA, Kutish GF, Nelson EA, Diel DG. Pathogenesis of Senecavirus A infection in finishing pigs. J Gen Virol. 2016;97:3267-3279.
5. Amass SF, Schneider JL, Miller CA, Shawky SA, Stevenson GW, Woodruff ME. Idiopathic vesicular disease in a swine herd in Indiana. J Swine Health Prod. 2004;12:192-196.
6. Pasma T, Davidson S, Shaw SL. Idiopathic vesicular disease in swine in Manitoba. Can Vet J. 2008;49:84-85.
7. Leme RA, Zotti E, Alcantara BK, Oliveira MV, Freitas LA, Alfieri AF, Alfieri AA. Senecavirus A: An emerging vesicular infection in Brazilian pig herds. Transbound Emerg Dis. 2015;62:603-611.
8. Vannucci FA, Linhares DC, Barcellos DE, Lam HC, Collins J, Marthaler D. Identification and complete genome of Seneca Valley virus in vesicular fluid and sera of pigs affected with idiopathic vesicular disease in Brazil. Transbound Emerg Dis. 2015;62:589-593.
9. Hause BM, Myers O, Duff J, Hesse RA. Senecavirus A in pigs, United States, 2015. Emerg Infect Dis. 2016;22:1323-1325.
10. Chen Z, Yuan F, Li Y, Shang P, Schroeder R, Lechtenberg K, Henningson J, Hause B, Bai J, Rowland RR, Clavijo A, Fang Y. Construction and characterization of a full-length cDNA infectious clone of emerging porcine Senecavirus A. Virology. 2016;497:111-124.
11. Farez S, Morley RS. Potential animal health hazards of pork and pork products. Rev Sci Tech. 1997;16:65-78.
*12. Houston E, Gimenez-Lirola LG, Magtoto R, Baum D, Pineyro PE. Seroprevalence of Senecavirus A in swine herds on the United States. Proc Am Assoc Vet Lab Diagn. San Diego, CA. 2017:128.
13. Resende TP, Marthaler DG, Vannucci FA. A novel RNA-based in situ hybridization to detect Seneca Valley virus in neonatal piglets and sows affected with vesicular disease. PLoS One. 2017:12:e0173190. doi:10.1371/journal.pone.0173190
14. Fukai K, Yamada M, Morioka K, Ohashi S, Yoshida K, Kitano R, Yamazoe R, Kanno T. Dose-dependent responses of pigs infected with foot-and-mouth disease virus O/JPN/2010 by the intranasal and intraoral routes. Arch Virol. 2015;160:129-139.
15. Yamada M, Fukai K, Morioka K, Nishi T, Yamazoe R, Kitano R, Shimada N, Yoshida K, Kanno T, Sakamoto K, Yamakawa M. Early pathogenesis of the foot-and-mouth disease virus O/JPN/2010 in experimentally infected pigs. J Vet Med Sci. 2018;80:689-700.
16. Mann JA, Hutchings GH. Swine vesicular disease: pathways of infection. J Hyg. 1980;84:355-363.
* Non-refereed reference.
